Isolation and expression analysis of glycerol-3-phosphate acyltransferase genes from peanuts (Arachis hypogaea L.)
X. Chia,b, Q. Yangc, L. Panb, N. Chenb, M. Chenb, T. Wangb, M. Wangb, Z. Yangb, X. Guand,* and S. Yub,*
aKey Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture, Oil Crops Research Institute, Chinese
Academy of Agricultural Sciences, Wuhan, 430062, P. R. China
bShandong Peanut Research Institute, Qingdao, 266100, P R China
cCollege of food science and engineering of Qingdao agricultural university, Qingdao, 266109, P R China
dSchool of Ocean Sciences, China University of Geosciences, Beijing 100083, P R China
*Corresponding authors: shanlinyu2012@163.com; guanxy@cugb.edu.cn
| |
SUMMARY
sn-Glycerol-3-phosphate acyltransferase (GPAT) catalyzes the committed step in the production of glycerolipids. The functions
of GPAT genes have been intensively studied in Arabidopsis, but not in peanuts (Arachis hypogaea L.). In this study, six AhGPAT genes were isolated from peanuts. Quantitative real-time RT-PCR analysis indicated that the AhGPAT9 transcript was more abundant in the stems, flowers, and seeds, whereas the transcript abundances of five other genes were
higher in the leaves or flowers than in the other tissues examined. During seed development, the transcript levels of AhGPAT9 gradually increased, whereas the transcript levels of the other five genes decreased. In addition, the levels of AhGPAT2 transcript were distinctly enhanced after exposure to all four kinds of stress treatments except for ABA-treated leaves.
The transcripts of AhGPAT1, AhGPAT6, AhGPAT8 and AhATS1 increased substantially in roots exposed to salt, drought, and ABA stress. The expressions of AhGPAT6, AhGPAT8, AhGPAT9 and AhATS1 were slightly higher in leaves under certain stress conditions than under normal conditions. The present study provides significant
information for modifying oil deposition and improving the abiotic stress resistance of peanuts through molecular breeding.
|
| |
RESUMEN
Aislamiento y análisis de la expresión del gen aciltransferasa glicerol-3-fosfato de cacahuete (Arachis hypogaea L.). La aciltransferasa sn-glicerol-3-fosfato (ATGP) cataliza el comprometido paso de la producción de glicerolípidos. Las funciones de los genes AhATGP se han estudiado intensivamente en Arabidopsis, pero no en cacahuete (Arachis hypogaea L.). En este estudio, seis genes AhATGP se aislaron a partir de cacahuetes. El análisis a tiempo real RT-PCR cuantitativa indicó que la transcripción AhATGP9 fue más abundante en tallos, flores y semillas, mientras que la abundancia de la transcripción de los otros cinco genes fueron
mayores en hojas o flores que en los otros tejidos examinados. Durante el desarrollo de la semilla, los niveles de transcripción
de AhATGP9 aumentaron gradualmente, mientras que los niveles de transcripción de otros cinco genes disminuyeron. Además, los niveles
de transcripción AhATGP2 mejoraron claramente después de la exposición a los cuatro tipos de tratamientos de estrés excepto para las hojas tratadas
con ABA. Las transcripciones de ATGP1, ATGP6, ATGP8 y AhATS1 aumentaron considerablemente en las raíces expuestas a sal, sequía y estrés de ABA. Las expresiones de AhGPAT6, AhGPAT8, AhGPAT9 y AhATS1 fueron ligeramente más altos en las hojas bajo ciertas condiciones de estrés que en condiciones normales. El presente estudio
proporciona información importante para utilizar en la modificación de la acumulación de aceite y mejorar la resistencia al
estrés abiótico de maní a través de mejoramiento molecular.
|
1. INTRODUCTIONTOP
Plant lipids are composed of a wide variety of fatty acids and their derivatives, including glycerolipids, lipid polyesters,
and sterols. Plant lipids are involved in a diverse range of metabolic reactions and play important physiological roles in
plant development, such as major components of cellular membranes, storage reserves, extracellular protective layers, and
signaling molecules (Chen et al., 2011a). The biosynthesis of these different types of lipids is controlled by a complex network of genes and proteins. sn-Glycerol-3-phosphate acyltransferases (GPAT) is the first enzyme in the pathway for the de novo synthesis of glycerolipids and is involved in different metabolic pathways and physiological processes (Yang et al., 2012). It catalyzes the transfer of an acyl group from acyl-coenzyme A (CoA) or acyl-acyl carrier protein (ACP) to the sn-1 position of sn-glycerol-3-phosphate (G3P). Plants contain three types of GPATs, which are located in plastids, mitochondria, and cytoplasm,
respectively (Xu et al., 2006; Li et al., 2011). The enzyme in plastids is soluble and uses acyl-ACP as the acyl donor, whereas the enzymes in the mitochondria and the
cytoplasm are bound to membranes and use acyl-CoA as the acyl donor (Murata and Tasaka, 1997).
In Arabidopsis, 10 genes have been identified as encoding GPAT enzymes located in various subcellular compartments, such as
plastids (AtATS1), mitochondria (AtGPAT1), and the endoplasmic reticulum (ER; AtGPAT8 and AtGPAT9) (Xu et al., 2006; Zheng et al., 2003; Gidda et al., 2009). The soluble, plastid-localized ATS1 (At1g32200) uses acyl-ACP substrates and exhibits sn-1 acyl transfer regio-specifically (Nishida et al., 1993). A second enzyme, GPAT9 (At5g60620), is not related to GPAT1–GPAT8 but is most homologous to the mammalian GPAT3, which
is directly involved in the synthesis of triacylglycerols in the adipose tissues (Cao et al., 2006). GPAT9 protein is localized to the ER (Gidda et al., 2009) and may be an acyl-CoA-dependent sn-1 GPAT that enables non-plastid glycerolipid synthesis. The remaining eight GPATs cluster together in a family (Zheng et al., 2003; Gidda et al., 2009; Beisson et al., 2007) which is not required for membrane or storage lipid biosynthesis. Instead, several members of the family clearly affect
the composition and quantity of cutin or suberin. They transfer acyl groups to the sn-2 position with three distinct clades which are associated with key stages in the morphological and functional evolution
of land plants and also coincide with a loss in phosphatase activity (Yang et al., 2012). Within the cutin-associated clade, GPAT4, GPAT6, and GPAT8 have been shown to behave as bifunctional sn-2 acyltransferase/phosphatase enzymes capable of generating 2-monoacylglycerol (MAG) products. They strongly prefer C16:0
and C18:1 ω-oxidized acyl-CoAs over unmodified or longer acyl chain substrates (Yang et al., 2012). In contrast, suberin-associated GPAT5 and GPAT7 possess sn-2 acyltransferase but not phosphatase activity, and can accommodate a broad chain-length range of ω-oxidized and unsubstituted
acyl-CoAs. The enzymes GPAT1–GPAT3 represent a distinct clade from the GPAT4/6/8 and the GPAT5/7 clades in the GPAT family
of Arabidopsis. Within this clade, phosphatase-minus GPAT1 can use dicarboxylic acyl-CoA substrates, whereas the same activity
could not be detected for GPAT2 and GPAT3. Even though GPAT2 and GPAT3 have lost their key amino acids in their phosphatase
domain, they retain their HXXXXD and CPEGT conserved acyl transferase domain motifs, and may thus be expected to function
as active acyltransferases (Yang et al., 2012).
In Arabidopsis, AtGPAT1 encodes a mitochondrial isozyme that is necessary for pollen development, although AtGPAT1 deficiency does not affect the levels of seed oil (Zheng et al., 2003). Analysis of loss-of-function mutants in Arabidopsis demonstrated an essential role of AtGPAT5 for suberin biosynthesis in the root and seed coat (Beisson et al., 2007). Monomer composition analysis and overexpression of AtGPAT5 in Arabidopsis and tobacco plants caused secretion of MAGs onto the surface of leaves (Li et al., 2007a). Similarly, AtGPAT4 and AtGPAT8 likely encode redundant activities necessary for the assembly of cutin monomers in the stems and leaves (Li et al., 2007b), whereas AtGPAT6 is involved in cutin assembly in sepals and petals (Li-Beisson et al., 2009). In Brassica napus, three homologous GPAT4 genes exhibited different expression patterns and distinct epigenetic features. A phenotypic rescue of a gpat4 gpat8 Arabidopsis double mutant and analysis of the gpat4 RNAi lines of B. napus suggested physiological roles of GPAT4s in cuticle formation of the rosette leaves, early flower development, pollen development, and the biosynthesis of storage
lipids (Chen et al., 2011b). Two homologous GPAT genes isolated from Echium pitardii have high similarity to the AtGPAT4/8 genes of Arabidopsis. Whereas the transcripts of EpGPAT1 were most abundant in seeds, roots, young leaves, and flowers, the transcripts of EpGPAT2 were most abundant in developing leaves and flowers. The ectopic expression of EpGPAT1 in the leaves of tobacco plants increased the levels of C16 and C18 hydroxyacids and a,ω-diacids in the cell wall fraction,
indicating a role for EpGPAT1 in the biosynthesis of cutin polyesters (Mañas-Fernández et al., 2010).
In vivo experiments showed that the overexpression of Arabidopsis AtAST1 in tobacco increased both the degree of unsaturation of fatty acids in phosphatidylglycerol (PG) and the resistance of tobacco
to chilling stress (Murata et al., 1992). An increase in the level of unsaturation of fatty acids in PG from rice plants transformed with an AtATS1 cDNA improved photosynthetic rates and growth at low temperatures (Ariizumi et al., 2002). The overexpression of LeATS1 increased the levels of PG cis-unsaturated fatty acids in the thylakoid membranes of tomato, which promoted recovery from chilling-induced photoinhibition
of photosystem I (PSI) (Sui et al., 2007). The increase in saturation of thylakoid membrane lipids in transgenic tobacco with expressed ATS1 from sweet pepper enhanced the thermotolerance of the photosynthetic apparatus of transgenic tobacco (Yan et al., 2008).
Given that the members of the GPAT family have several complicated roles during plant development and acclimation to stressful
conditions, functional analyses of each member of the gene family should be helpful in elucidating the roles of GPAT isoforms.
The peanut (Arachis hypogaea L.) is an allotetraploid species (2n=4×=40, AABB) and one of the five most important oilseed crops worldwide. It is grown
extensively in tropical, subtropical, and temperate climates. The peanut seed comprises around 50% oil, of which approximately
80% consists of oleic (36–67%) and linoleic (15–43%) acids (Chi et al., 2011). Several molecular studies of lipid biosynthesis in peanuts have been reported in recent years. However, there have been
no reports about the function of the GPAT family proteins in peanuts. In the present study, we isolated six novel GPAT genes from peanuts. The expression patterns of these genes were investigated in different tissues and at different stages
of seed development. Expressions of these genes were also analyzed under conditions of cold, salt, drought, and ABA stress.
Our findings should be of value in efforts to modify lipid biosynthesis in peanut seeds and to provide a theoretical basis
for the study of abiotic stress tolerance in peanut.
2. MATERIALS AND METHODSTOP
2.1. Plant materialsTOP
Peanut plants (A. hypogaea L. cultivar Huayu 19) were grown in a growth chamber with a 16 h light/8 h dark photoperiod at 26 °C/22 °C day/night temperatures.
Leaves, stems, cotyledons, hypocotyls, and roots were sampled from the seedlings at the trefoil leaf stage. Seeds were sampled
at 10, 20, 30, 40, 50, and 60 days after pegging (DAP). Flowers were collected when the seedlings were in the flowering phase.
For the cold treatment, seedlings in the soil at the trefoil leaf stage were kept at 4 °C, and leaves were sampled separately
either before cold treatment (0 h) or after continuous exposure to 4 °C for 1, 3, 6, 12, 24, 48, or 72 h. For stress treatments,
the roots of seedlings grown in soil were flushed carefully with tap water to remove all soil, and then submerged in solutions
of 200 mM NaCl, 20% PEG-6000, or 100 μM ABA. Leaves and roots were sampled separately after treatment for 0, 1, 3, 6, 12,
24, 48, or 72 h. All samples were immediately frozen in liquid nitrogen and stored at −80 °C until required.
2.2. Identification of glycerol-3-phosphate acyltransferase family genes in a peanut cDNA library using bioedit softwareTOP
The cDNA sequences used in this study came from three cDNA libraries from three institutes (data not shown): Shandong Peanut
Research Institute, Oil Crops Research Institute of The Chinese Academy of Agricultural Sciences, and Crops Research Institute
of Guangdong Academy of Agricultural Sciences. All expressed sequence tags (ESTs) of the 36,741 cDNA sequences were saved
as FASTA format. The amino acid sequences of glycerol-3-phosphate acyltransferase genes of Arabidopsis, AtGPAT2 (NP_563651), AtGPAT9 (NP_568925) and AtATS1 (NP_174499) were used to search for homogeneous genes from the peanut cDNA library. Before searching for members of the GPAT gene family, a local nucleotide database file was created using Bioedi software. A local BLAST procedure was then run to
find the homologous genes of the GPAT family. Using this method, we found six genes that may encode GPAT proteins.
2.3. Total RNA isolation and cDNA synthesisTOP
The total RNA was extracted using the RNeasy Plant Mini kit (Qiagen, Valencia, CA, USA). Contamination with genomic DNA was
eliminated by treatment with recombinant DNase I (Qiagen), as recommended by the vendor. Only RNA preparations having an A260/A280
ratio of 1.8–2.0 and an A260/A230 ratio >2.0 were used for subsequent analysis. The integrity of RNA was verified by electrophoresis
through 2% agarose gels, followed by SYBR Green staining. First-strand cDNA synthesis was carried out with 2 μg RNA using
an RT-PCR kit (Promega, WI, USA) according to the manufacturer’s procedure.
2.4. Isolation of full-length cDNA sequencesTOP
We performed PCR with the LA PCR system (TaKaRa) using 2.5 μL of 10×PCR buffer with MgCl2, 1 μL of each primer (10 μM), 4.0 μL of 10 mM dNTPs, 1 μL of cDNA sample, 0.5 μL of LA Taq™ DNA polymerase, and 15 μL of
double-distilled water. The PCR products were separated by electrophoresis through a 1% agarose gel, and purified using a
Gel Extraction Kit (Takara) according to the manufacturer’s protocol. The purified products were then cloned into the pMD18-T
Easy vector (Takara) and sequenced (Shangon, Shanghai).
2.5. Sequence analysisTOP
The open reading frames (ORFs) and encoded amino acid sequences of all the genes were deduced using BioXM 2.6. The physicochemical
properties of the deduced protein were predicted using Protparam (http://www.expasy.ch/tools/protparam.html). Active sites of the protein sequence were analyzed by comparison against the PROSITE database. Predicted transmembrane
domain (TMDs) in GPAT proteins were identified using the TMHMM Server (version 2.0) (http://www.cbs.dtu.dk/services/TMHMM) and visual inspection. The putative subcellular localizations of the candidate proteins were estimated by TargetP (http://www.cbs.dtu.dk/services/TargetP/) and Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html).
2.6. Gene structure prediction and conserved motif scanningTOP
The gene structure display server (GSDS) program (Guo et al., 2007) was used to illustrate exon/intron organization for individual desaturase genes by comparison of the cDNAs with their
corresponding genomic DNA sequences. To identify the conserved motifs, MEME (Multiple Expectation Maximization for Motif Elicitation)
version 4.9.1 (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) was employed with a set of parameters as follows: number of repetitions – any, maximum number of motifs –20, optimum motif
width set to ≥6 and ≤200 (Bailey and Elkan 1995). The motifs obtained were recorded using the SMART (http://smart.embl-heidelberg.de/) and NCBI-CDD (National Center for Biotechnology Information Conserved Domain Database) search programs.
2.7. Phylogenetic analysisTOP
Homologs of each member of the Arabidopsis GPAT family were identified by BLASTP searches with datasets from Phytozome v9.1 (www.phytozome.net). Only those sequences with an e-value less than e−50 were considered as members of the GPAT family. In each tree, gene sequences other than Arabidopsis and peanut GPATs were
displayed using the nomenclature with the following abbreviations: Ah, Arachis hypogaea; At, Arabidopsis thaliana; Glyma, Glycine max; Medtr, Medicago truncatula; Pp, Physcomitrella patens; Cre, Chlamydomonas reinhardtii; Vocar, Volvox carteri. Table 1 provides a detailed description of the proteins used and the corresponding accession numbers. Amino acid sequences were aligned
using the ClustalX program with the implanted BioEdit (Thompson et al., 1994). The neighbor-joining (NJ) method in MEGA4 (Tamura et al., 2007) was used to construct the phylogenetic tree. Bootstrapping with 1,000 replicates was used to establish the confidence limits
of the tree branches. Default program parameters were used. Bootstrap values from the neighbor-joining analyses were listed
to the left of each node, and values higher than 50 were shown.
Table 1. The GPAT enzymes used for the phylogenetic analyses
| Kingdom |
Specie |
Taxa terminologies |
Gene symbol |
Database |
Access |
Length (aa) |
| Viridiplantae |
Arabidopsis thaliana |
At |
ATS1 |
JGI |
AT1G32200.2 |
459 |
|
|
|
GPAT1 |
JGI |
AT1G06520.1 |
585 |
|
|
|
GPAT2 |
JGI |
AT1G02390.1 |
530 |
|
|
|
GPAT3 |
JGI |
AT4G01950.1 |
520 |
|
|
|
GPAT4 |
JGI |
AT1G01610.1 |
503 |
|
|
|
GPAT5 |
JGI |
AT3G11430.1 |
502 |
|
|
|
GPAT6 |
JGI |
AT2G38110.1 |
501 |
|
|
|
GPAT7 |
JGI |
AT5G06090.1 |
500 |
|
|
|
GPAT8 |
JGI |
AT4G00400.1 |
500 |
|
|
|
GPAT9 |
JGI |
AT5G60620.1 |
376 |
|
Glycine max |
Glyma |
ATS1 |
JGI |
Glyma01g01800.1 |
253 |
|
|
|
ATS1 |
JGI |
Glyma09g34110.1 |
470 |
|
|
|
GPAT1 |
JGI |
Glyma02g45600.1 |
539 |
|
|
|
GPAT1 |
JGI |
Glyma08g42210.1 |
552 |
|
|
|
GPAT1 |
JGI |
Glyma14g03210.1 |
540 |
|
|
|
GPAT1 |
JGI |
Glyma18g12750.1 |
527 |
|
|
|
GPAT2 |
JGI |
Glyma03g37970.1 |
522 |
|
|
|
GPAT2 |
JGI |
Glyma03g37990.1 |
481 |
|
|
|
GPAT2 |
JGI |
Glyma02g01400.1 |
555 |
|
|
|
GPAT2 |
JGI |
Glyma10g01420.1 |
553 |
|
|
|
GPAT2 |
JGI |
Glyma19g40590.1 |
537 |
|
|
|
GPAT3 |
JGI |
Glyma14g33830.1 |
417 |
|
|
|
GPAT3 |
JGI |
Glyma14g33860.1 |
534 |
|
|
|
GPAT3 |
JGI |
Glyma13g02250.1 |
446 |
|
|
|
GPAT4 |
JGI |
Glyma07g07580.1 |
499 |
|
|
|
GPAT4 |
JGI |
Glyma03g01070.1 |
500 |
|
|
|
GPAT5 |
JGI |
Glyma02g41660.1 |
467 |
|
|
|
GPAT5 |
JGI |
Glyma14g07290.1 |
512 |
|
|
|
GPAT6 |
JGI |
Glyma01g27900.1 |
492 |
|
|
|
GPAT6 |
JGI |
Glyma18g42580.1 |
539 |
|
|
|
GPAT6 |
JGI |
Glyma20g16980.1 |
501 |
|
|
|
GPAT6 |
JGI |
Glyma10g23560.1 |
489 |
|
|
|
GPAT6 |
JGI |
Glyma03g14180.1 |
362 |
|
|
|
GPAT6 |
JGI |
Glyma07g17720.1 |
496 |
|
|
|
GPAT9 |
JGI |
Glyma05g26140.1 |
238 |
|
|
|
GPAT9 |
JGI |
Glyma08g09080.1 |
373 |
|
|
|
GPAT9 |
JGI |
Glyma09g21150.1 |
376 |
|
Medicago truncatula |
Medtr |
ATS1 |
JGI |
Medtr5g029110.1 |
457 |
|
|
|
GPAT1 |
JGI |
Medtr5g098930.1 |
537 |
|
|
|
GPAT1 |
JGI |
Medtr3g062190.1 |
277 |
|
|
|
GPAT2 |
JGI |
Medtr1g106370.1 |
542 |
|
|
|
GPAT4 |
JGI |
Medtr8g031940.1 |
505 |
|
|
|
GPAT5 |
JGI |
Medtr5g087710.1 |
523 |
|
|
|
GPAT6 |
JGI |
Medtr3g024620.1 |
496 |
|
|
|
GPAT6 |
JGI |
Medtr1g059560.1 |
504 |
|
|
|
GPAT9 |
JGI |
Medtr8g129160.1 |
371 |
|
Arachis hypogaea |
Ah |
ATS1 |
NCBI |
KC762933 |
451 |
|
|
|
GPAT1 |
NCBI |
JN032676 |
555 |
|
|
|
GPAT2 |
NCBI |
HQ589243 |
544 |
|
|
|
GPAT6 |
NCBI |
HQ589244 |
499 |
|
|
|
GPAT8 |
NCBI |
JX843442 |
505 |
|
|
|
GPAT9 |
NCBI |
JX843441 |
376 |
|
Physcomitrella patens
|
Pp |
ATS1 |
JGI |
Pp1s136_120V6.1 |
494 |
|
|
|
GPAT6 |
JGI |
Pp1s9_453V6.1 |
510 |
|
|
|
GPAT6 |
JGI |
Pp1s134_51V6.1 |
504 |
|
|
|
GPAT6 |
JGI |
Pp1s72_49V6.1 |
516 |
|
|
|
GPAT6 |
JGI |
Pp1s117_125V6.1 |
517 |
|
|
|
GPAT6 |
JGI |
Pp1s42_150V6.1 |
513 |
|
|
|
GPAT6 |
JGI |
Pp1s281_69V6.1 |
529 |
|
|
|
GPAT6 |
JGI |
Pp1s117_135V6.1 |
538 |
|
|
|
GPAT9 |
JGI |
Pp1s150_100V6.1 |
276 |
|
|
|
GPAT9 |
JGI |
Pp1s138_27V6.1 |
389 |
|
Chlamydomonas reinhardtii |
Cre |
ATS1 |
JGI |
Cre02g143000.t1.2 |
410 |
|
|
|
GPAT9 |
JGI |
Creg6130.t1 |
456 |
|
Volvox carteri |
Vocar |
ATS1 |
JGI |
Vocar20013783m |
406 |
|
|
|
GPAT9 |
JGI |
Vocar20002974m |
435 |
2.8. Quantitative real-time RT-PCRTOP
A quantitative Real-time RT-PCR (qRT-PCR) analysis was performed using a LightCycler 2.0 instrument system (Roche, Germany).
The alpha tubulin 5 gene (AhTUA5) was taken as a reference gene (Chi et al., 2012). Seven pairs of gene-specific primers (Table 2) were designed after analysis of the sequences of target genes. qRT-PCR reactions were performed using the SYBR Premix Ex
Taq polymerase (TaKaRa, Japan) according to the manufacturer’s instructions. Each 20-μL reaction comprised 2 μL of template,
10 μL of 2× SYBR Premix, and 0.4 μL (200 nM) of each primer. The reactions were subjected to an initial denaturation step
of 95 °C·10 s−1, followed by 40 cycles of 95 °C·5s−1, 60 °C·30s−1 and 72 °C·10s−1. A melting curve analysis was performed at the end of the PCR run over the range 60–95 °C, increasing the temperature stepwise
by 0.5 °C every 10 s. The baseline and quantification cycle (CP) were automatically determined using the Light Cycler Software.
Zero template controls were included for each primer pair, and each PCR reaction was carried out in triplicate. The relative
quantification method (delta-delta Cp) was used to evaluate quantitative variation (Livak and Schmittgen, 2001).
Table 2. DNA sequences of oligonucleotide primers used in this study
| Name |
Oligonucleotide sequence 5’–3’ |
| Full-length cDNA sequence cloning |
|
| ATS1-F |
ATGAACGGGTCTCTCGCTCA |
| ATS1-R |
CTAGTTCCACGGCTGTGACAA |
| GPAT1-F |
ATGGTGTTTCCAATGGTGCT |
| GPAT1-R |
TCACGACAGCAAAGTTTCTC |
| GPAT2-F |
ATGGCTAAAATGTTCAGAGCT |
| GPAT2-R |
CTAAGATTTACCACACGCTC |
| GPAT6-F |
ATGGTCATGGGAGCCTTTTC |
| GPAT6-R |
TTAAGCTTTGTTCTCCTTGTTAG |
| GPAT8-F |
ATGGCAGCGCCGAAACCGA |
| GPAT8-R |
TCACTTCTTGGAACTGTACATGG |
| GPAT9-F |
ATGATGAGGAAGACCAATCC |
| GPAT9-R |
TTACTTTTCTTCCAAGCGCC |
| Real-time RT-PCR |
|
| qTUA5-F |
CTGATGTCGCTGTGCTCTTGG |
| qTUA5-R |
CTGTTGAGGTTGGTGTAGGTAGG |
| qATS1-F |
TTCCGTGACTGAGCAATATACTGTG |
| qATS1-R |
GGCTGTGACAACGAGACTTTAGG |
| qGPAT1-F |
CCTACTTCACTGGCTTTGTCTCTG |
| qGPAT1-R |
CATTGGGCTTGGATTGTTCACC |
| qGPAT2-F |
GGTGTCAGAAGCAGAGAAGAGAAG |
| qGPAT2-R |
TGGCGAGGATTAGGGCATAGG |
| qGPAT6-F |
GCTTCCCTCTTAACCTTCCTATGG |
| qGPAT6-R |
TCCGCTTTGCCCTTTCTTTGG |
| qGPAT8-F |
TCACCTACTCCGTCAGCAAGC |
| qGPAT8-R |
GCATTGAACCGCAGCAAGAAG |
| qGPAT9-F |
AACCTAACATTGAAGATTACCT |
| qGPAT9-R |
ATTGACTTGAAGCACCTTAA |
2.9. Seed lipid analysisTOP
Lipid content in seed was determined by a standard Soxhlet extraction method (Harwood, 1984). From each cultivar, 1 g sample was ground and then extracted with petroleum ether in a Soxhlet apparatus for 8 h. Petroleum
ether was then volatilized in the draft. The experiment was carried out in triplicate. Lipid content was expressed as % of
seed dry weight.
3. RESULTSTOP
3.1. Isolation of glycerol-3-phosphate acyltransferase genes from peanutsTOP
Six genes that likely encode glycerol-3-phosphate acyltransferase (GPAT) proteins were found using Bioedit software. They
were cloned and designated as AhATS1, AhGPAT1, AhGPAT2, AhGPAT6, AhGPAT8, and AhGAPT9 according to the homologous genes identified in Arabidopsis. Among the six genes, two genes have the complete open reading
frame (ORF) in the peanut cDNA library and cloned by conventional RT-PCR, whereas four genes were cloned using the rapid amplification
of cDNA ends (RACE) method. The ORFs of the five genes were 1,356 bp, 1,668 bp, 1,635 bp, 1,500 bp, 1,518 bp, and 1,131bp
in length, encoding 451, 555, 544, 499, 505, and 376 amino acids, respectively. The genomic sequences were 5,766 bp, 2,146
bp, 2,209 bp, 3,176 bp, 4,474bp, 4,970 bp in length, respectively (Table 3). The sequence information of six genes was submitted to Genbank, with the Genbank identification numbers KC762933, JN032676,
HQ589243, HQ589244, JX843442, and JX843441, respectively.
Table 3. Glycerol-3-phosphate acyltransferase genes in peanuts
| Protein |
Accession |
Len (aa) |
ORF (bp) |
5’ upstream region (bp) |
3’ downstream region (bp) |
Genomic sequences (bp) |
Molecular mass (kDa) |
PI |
| ATS1 |
KC762933 |
451 |
1356 |
89 |
496 |
5766 |
49.5908 |
9.14 |
| GPAT1 |
JN032676 |
555 |
1668 |
78 |
137 |
2146 |
62.6046 |
9.05 |
| GPAT2 |
HQ589243 |
544 |
1635 |
62 |
61 |
2209 |
61.8657 |
9.34 |
| GPAT6 |
HQ589244 |
499 |
1500 |
113 |
10 |
3176 |
55.5391 |
9.19 |
| GPAT8 |
JX843442 |
505 |
1518 |
96 |
316 |
4474 |
51.642 |
9.09 |
| GPAT9 |
JX843441 |
376 |
1131 |
136 |
158 |
4970 |
43.5418 |
9.09 |
A search using NCBI BLAST revealed that six GPAT proteins have high sequence similarities with GPATs in Arabidopsis. AhATS1
shares 55.1% sequence similarity with AtATS1. AhGPAT1 shows 55.3% sequence similarity with AtGPAT1, AhGPAT6 shares 78.2% similarity with AtGPAT6. The AhGPAT2 protein shares 52.8% and 52.0% sequence similarity with AtGPAT2 and AtGPAT3, respectively, and AhGPAT9 shows 79% similarity with AtGPAT9. The AhGPAT8 protein is most similar to AtGPAT4 (77.9%), and AtGPAT8 (78.4%), both of which have been implicated in the synthesis
of cutin polymers (Li et al., 2007b).
As shown in Figures 1 and 2, alignment of the deduced polypeptide sequences of six GPAT proteins demonstrates that they are similar in length and share
several features that are characteristic of other plastidial and membrane-bound GPATs from evolutionarily diverse organisms.
These features include the presence of four conserved amino acid motifs (AT-I to AT-IV) which are important for acyltransferase
activity (Mañas-Fernández et al., 2010). The typical acyltransferase (AT) domain is localized within the C-terminal half of the molecule. Residues implicated in
catalysis, such as histidine and aspartic acid residues in AT-I, glycine residues in AT-III, and a proline residue in AT-IV
are all present in peanut GPATs, as are the arginine (AT-II) and glutamic (serine) (AT-III) residues involved in binding to
the G3P substrate (Gonzalez-Baró et al., 2007). In addition to the AT region, a haloacid dehalogenase (HAD)-like domain is found in the N-terminal half of AhGPAT6 and AhGPAT8. This conserved domain is present in a super-family of proteins, most of which are phosphohydrolases. Close inspection of
this region in AhGPAT6 and AhGPAT8 and their putative orthologues reveal the presence of highly conserved motifs, named HAD-I through HAD-IV (Figure 1), which have been described in HAD-like proteins (Burroughs et al., 2006). They include the typical DXD signature (which contains critical aspartic acid residues that act as a nucleophile during
catalysis), the extremely conserved threorine and lysin residues in HAD-II and HAD-III boxes, respectively (both of which
contribute to the stability of the reaction intermediates), and a GDXXXD motif in HAT-IV that contains acidic residues required
for coordination to the Mg2+ ion in the active site.
|
 Figure 1. Amino acid alignment of peanut plastidial ATS1 proteins and closely related proteins found in the GenBank. Residues shared
by a fraction of sequences above 0.5 were shaded, identical residues in black, similar residues in grey. AT-like domains were
boxed (ATI to AT-IV). Critical residues previously identified in similar proteins were marked by dots (binding site in AT
domain) or triangles (catalytic residues in AT domain). GenBank accession numbers were as follows: Arachis hypogaea (AhATS1, KC762933), Arabidopsis thaliana (AtATS1, NP_174499), Glycine max (GmATS1, XP_003516958), Physcomitrella patens (PpATS1, XP_001771299), Medicago truncatula (MeATS1, XP_003612801), Chlamydomonas reinhardtii (CrATS1, XP_001694977), Volvox carteri (VoATS1, XP_002950506). Figure 1. Amino acid alignment of peanut plastidial ATS1 proteins and closely related proteins found in the GenBank. Residues shared
by a fraction of sequences above 0.5 were shaded, identical residues in black, similar residues in grey. AT-like domains were
boxed (ATI to AT-IV). Critical residues previously identified in similar proteins were marked by dots (binding site in AT
domain) or triangles (catalytic residues in AT domain). GenBank accession numbers were as follows: Arachis hypogaea (AhATS1, KC762933), Arabidopsis thaliana (AtATS1, NP_174499), Glycine max (GmATS1, XP_003516958), Physcomitrella patens (PpATS1, XP_001771299), Medicago truncatula (MeATS1, XP_003612801), Chlamydomonas reinhardtii (CrATS1, XP_001694977), Volvox carteri (VoATS1, XP_002950506).
|
|
|
 Figure 2. Amino acid alignment of peanut membrane-bound GPAT proteins and closely related proteins found in the GenBank. Residues shared
by a fraction of sequences above 0.5 were shaded, identical residues in black, similar residues in grey. Putative trans membrane
domains of peanut GPAT proteins were underlined. AT and HAD-like domains were boxed (ATI to AT-IV) or marked by lines (HAD-I
to HAD-IV), respectively. Critical residues previously identified in similar proteins were marked by asterisks (HAD domain),
dots (binding site in AT domain) or triangles (catalytic residues in AT domain). GenBank accession numbers were as follows:
Arachis hypogaea (AhGPAT1, JN032676; AhGPAT2, HQ589243; AhGPAT6, HQ589244; AhGPAT8, JX843442; AhGPAT9, JX843441), Arabidopsis thaliana (AtGPAT1, NP_563768; AtGPAT2, NP_563651; AtGPAT3, NP_192104; AtGPAT4, NP_171667; AtGPAT5, NP_187750; AtGPAT6, NP_181346;
AtGPAT7, NP_196227; AtGPAT8, NP_191950; AtGPAT9, NP_568925), Glycine max (GmGPAT1, XP_003545142; GmGPAT2, XP_003520759; GmGPAT3, XP_003536864; GmGPAT6, XP_003529144; GmGPAT8, XP_003520970; GmGPAT9,
XP_003533946). Figure 2. Amino acid alignment of peanut membrane-bound GPAT proteins and closely related proteins found in the GenBank. Residues shared
by a fraction of sequences above 0.5 were shaded, identical residues in black, similar residues in grey. Putative trans membrane
domains of peanut GPAT proteins were underlined. AT and HAD-like domains were boxed (ATI to AT-IV) or marked by lines (HAD-I
to HAD-IV), respectively. Critical residues previously identified in similar proteins were marked by asterisks (HAD domain),
dots (binding site in AT domain) or triangles (catalytic residues in AT domain). GenBank accession numbers were as follows:
Arachis hypogaea (AhGPAT1, JN032676; AhGPAT2, HQ589243; AhGPAT6, HQ589244; AhGPAT8, JX843442; AhGPAT9, JX843441), Arabidopsis thaliana (AtGPAT1, NP_563768; AtGPAT2, NP_563651; AtGPAT3, NP_192104; AtGPAT4, NP_171667; AtGPAT5, NP_187750; AtGPAT6, NP_181346;
AtGPAT7, NP_196227; AtGPAT8, NP_191950; AtGPAT9, NP_568925), Glycine max (GmGPAT1, XP_003545142; GmGPAT2, XP_003520759; GmGPAT3, XP_003536864; GmGPAT6, XP_003529144; GmGPAT8, XP_003520970; GmGPAT9,
XP_003533946).
|
|
The AhATS1 protein is probably located in chloroplast, as predicted using the TargetP Server and Predotar tools. The N-terminal end
of AhATS1 had a high proportion of hydroxylated and small, hydrophobic amino acids, which is typical of a chloroplast transit peptide.
The AhGPAT1 and AhGPAT2 proteins also possess an extended N-terminal region that exhibits characteristics of a mitochondrial targeting peptide. All
of the other GPAT proteins lack any recognizable N-terminal intracellular targeting signal motifs, but do contain putative
C-terminal ER retrieval signals.
3.2. Gene structures and distribution of conserved motifsTOP
Genes in the same clade had more similar exon/intron structures than those genes in the other clades (Figure 3). Both ATS1 and GPAT9 clade members had twelve exons, whereas the GPAT4/8 clade members possessed four exons. All of the
remaining six GPAT clade members had two exons, except for AhGPAT1, which possessed three exons.
|
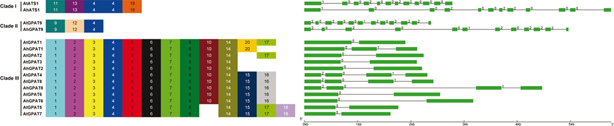 Figure 3. The conserved motifs and exon/intron structures of the peanut and Arabidopsis GPAT genes. Schematic representation of motifs
identified in peanut GPAT proteins using MEME motif search tool. Each motif was represented by a number in a colored box.
Length of box did not correspond to length of motif. Boxes represented the exons and lines represented introns. The sizes
of exons and introns could be estimated using the scale at the bottom. The numbers above the boxes and lines indicated the
splicing phases of the GPAT sequences, 0 referred to phase 0,1 to phase 1, and 2 to phase 2. Figure 3. The conserved motifs and exon/intron structures of the peanut and Arabidopsis GPAT genes. Schematic representation of motifs
identified in peanut GPAT proteins using MEME motif search tool. Each motif was represented by a number in a colored box.
Length of box did not correspond to length of motif. Boxes represented the exons and lines represented introns. The sizes
of exons and introns could be estimated using the scale at the bottom. The numbers above the boxes and lines indicated the
splicing phases of the GPAT sequences, 0 referred to phase 0,1 to phase 1, and 2 to phase 2.
|
|
The MEME motif search tool was employed to identify the conserved motifs present in peanuts and Arabidopsis GPAT proteins (Figure 3), and 20 distinct motifs were identified. Most of the motifs belonged to the regions that represented the typical domains
of acyltransferase. The motif 4 was found in all the members of the GPAT family proteins. Both of the ATS1 proteins had the
motifs 4, 11, 13, and 19, whereas the GPAT9 proteins all possessed the motifs 4, 9, and 12. The conserved motifs 1–8 and 14
were present in all of the remaining eight GPAT clade members. Both of the GPAT1 proteins had the motifs 1–8, 10, 14, and
20, whereas another motif 17 was present in AtGPAT1. All proteins belonging to GPAT2/3 clade had the motifs 1–8, 10, and 14,
except for AtGPAT2, which had another motif 17. All GPAT4–GPAT8 clade members possessed the motifs 1–8, 14, and 15. The motifs
10 and 16 were present in GPAT4/6/8 proteins, whereas the motifs 17 and 18 existed in GPAT5/7 proteins.
3.3. Phylogenetic analysisTOP
To examine the relationships among different sources of GPAT genes, the neighbor-joining method was used to construct phylogenetic
trees and all tree topologies were highly congruent (Figure 4). As shown in the phylogenetic tree, all of the GPATs fell into three distinct clades: the ATS1 clade, GPAT9 clade, and GPAT1–GPAT8
clades.
|
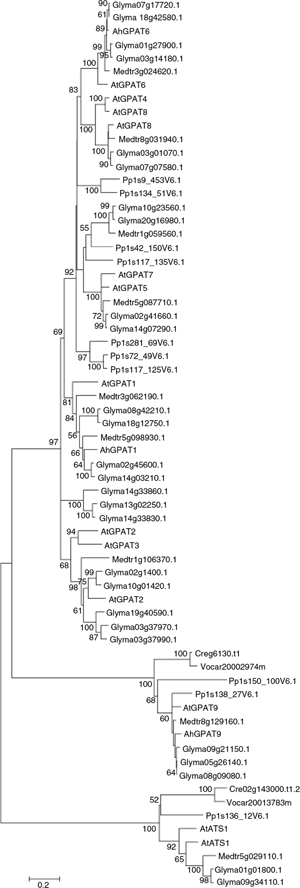 Figure 4. Neighbor-joining tree based on the deduced amino acid sequences of GPATs. Gene sequences other than Arabidopsis and peanut
GPATs were shown by their nomenclatures found at www.phytozome.org, with the abbreviations. Bootstrap values from neighbor-joining analyses were listed to the left of each node, with values
higher than 50 shown. Figure 4. Neighbor-joining tree based on the deduced amino acid sequences of GPATs. Gene sequences other than Arabidopsis and peanut
GPATs were shown by their nomenclatures found at www.phytozome.org, with the abbreviations. Bootstrap values from neighbor-joining analyses were listed to the left of each node, with values
higher than 50 shown.
|
|
Searches against prokaryote and non-photosynthetic eukaryotic sequences, and of the fully sequenced genomes of Chlamydomonas, Volvox and other algae do not identify any GPATs with significant similarities (BlastX E <10−5) to the sn-2 GPATs found in land plants (Yang et al., 2012). In contrast, plastid-localized ATS1 and GPAT9 were found in the algal genomes. Thus the sn-2 GPAT family clearly belongs to a lineage specific to land plants and evolved to provide pathways for functions not present
in other organisms. The AhATS1 protein was grouped with ATS1 enzymes from higher plants and green algae, and lie apart from
membrane-bound GPAT clades. AhGPAT9 clustered with GPAT9 from higher plants and green algae, apart from the subgroup comprised
of GPAT1–GPAT8 from higher plants. The sn-2 GPAT family also fell into three distinct conserved subfamilies. It was assumed that the GPAT4/6/8 clade is the most ancient
and arose early during the evolution of land plants (bryophytes), which is involved in the assembly of cutin or cutin-like
polymers in the first land plants (Yang et al., 2012). In contrast, the phosphatase-minus GPAT1-3 and 5/7 clades diverged later with the appearance of tracheophytes (Yang et al., 2012). Whereas AhGPAT8 was grouped with GPAT4 and GPAT8 from higher plants, AhGPAT6 fell into the GPAT6 subfamily. Sequences of
the GPAT1–GPAT3 clade were more divergent compared with the GPAT4/6/8 and GPAT5/7 clades. AhGPAT1 and AhGPAT2 were grouped
with their respective GPAT1 or GPAT2/3 enzymes from higher plants, and lie apart from the GPAT4–GPAT8 clades.
3.4. Tissue-specific expression patternsTOP
Quantitative real-time RT-PCR (qRT-PCR) was employed to confirm the expression patterns of the six novel genes in different
peanut tissues and at different stages of seed development. The alpha tubulin 5 (AhTUA5) gene was used as an internal reference control for total RNA input (Chi et al., 2012). As shown in Figure 5, these six genes displayed specific temporal and spatial expression patterns across different tissues and developmental stages.
AhATS1 showed higher transcript abundance in flowers and leaves than in any of the other tissues tested. The highest abundance of
AhGPAT1 transcript was in leaves and the lowest was in stems and flowers. Levels of AhGPAT2 transcript were highest in leaves, followed by stems and seeds, with the lowest levels in roots and flowers. AhGPAT6 and AhGPAT8 had similar expression patterns, showing higher transcript abundance in leaves and roots. AhGPAT9 exhibited its highest transcript accumulation in stems followed by flowers and seeds.
|
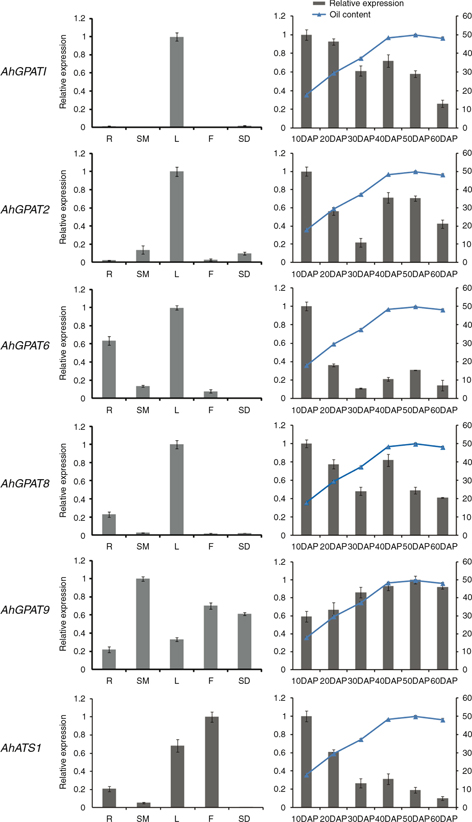 Figure 5. Expression analysis of six AhGPAT genes using qRT-PCR in five peanut tissues and at six stages of seed development. R, root; SM, stem; L, leaf; F, flower;
SD, seed. The relative mRNA abundance was normalized with respect to the peanut AhTUA5 gene. The bars were standard deviations (SD) of three technical repetitions. Figure 5. Expression analysis of six AhGPAT genes using qRT-PCR in five peanut tissues and at six stages of seed development. R, root; SM, stem; L, leaf; F, flower;
SD, seed. The relative mRNA abundance was normalized with respect to the peanut AhTUA5 gene. The bars were standard deviations (SD) of three technical repetitions.
|
|
The expression patterns of six GPAT genes across six developmental stages of seeds are also shown in Figure 5. Levels of AhGPAT1 transcript were maximal at 10 days after pegging (DAP) and decreased gradually thereafter. The expression patterns of AhGPAT2 and AhGPAT6 were similar over the course of seed development, with higher levels of AhGPAT2 and AhGPAT8 transcripts seen at 10 DAP and 40 DAP. The expression levels of AhATS1 and AhGPAT6 were highest at the initial stage of seed development but dramatically decreased in abundance during later stages. The AhGPAT9 transcript remained relatively low at the initial stage of seed development but increased gradually during later stages of
seed development. In peanut cultivar Huayu19, seed lipid content was low in the first period of lipid accumulation, but was
characterized by a drastic increase during the initial four stages after pegging (Figure 5). The seed lipid content reached a maximum value of 49.75% at 50 DAP and decreased thereafter at 60 DAP. The expressions
of the AhGPAT9 gene coincided with the lipid accumulation rate in peanut seed, whereas the expressions of other AhGPAT genes were not in complete agreement with seed lipid accumulation rate, especially in the earlier stages of seed development
like the period from 10 to 30 DAP. These results indicated that AhGPAT9 may be an important component in the lipid biosynthesis process.
3.5. Expression patterns of AhGPATs in peanut under abiotic stressTOP
To confirm the expression patterns of six GPAT genes under cold, salt, drought and ABA stress, we monitored the changes in these transcripts in peanut leaves and roots.
Figure 6 shows the expression patterns of six GPAT genes in peanut leaves upon cold treatment. Transcript levels of AhGPAT1 in the leaves decreased distinctly and rapidly between 1 h and 6 h after cold treatment, and increased thereafter. The levels
of AhGPAT2 transcript gradually accumulated between 1 h and 24 h after cold treatment, and then decreased drastically, with a peak level
of about 4-fold increase at 24 h. The expressions of AhGPAT6 and AhGPAT8 were slightly increased under cold stress, with a peak level at 1 h, and then decreased gradually. The expression of AhATS1 and AhGPAT9 gradually decreased under cold stress, while the lowest level was detected at 72 h.
|
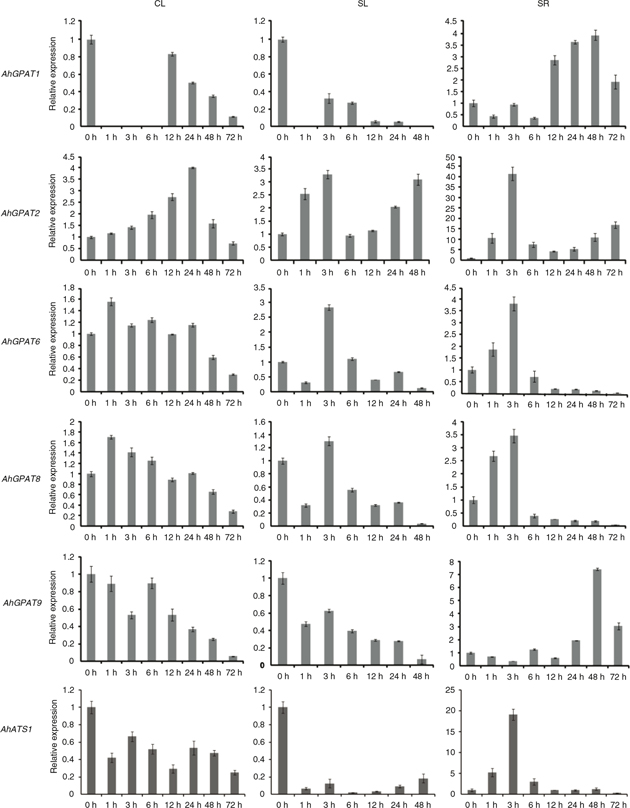 Figure 6. Expression analysis of six AhGPAT genes using qRT-PCR under cold and salt stress. The relative mRNA abundance was normalized with respect to the peanut AhTUA5 gene. The bars were standard deviations (SD) of three technical repetitions. CL (0 h to 72 h), leaves exposed to cold (4
°C) treatment; SL (0 h to 48 h), leaves exposed to high salt (200 mM NaCl) treatment; SR (0 h to 72 h), roots exposed to high
salt (200 mM NaCl) treatment. Figure 6. Expression analysis of six AhGPAT genes using qRT-PCR under cold and salt stress. The relative mRNA abundance was normalized with respect to the peanut AhTUA5 gene. The bars were standard deviations (SD) of three technical repetitions. CL (0 h to 72 h), leaves exposed to cold (4
°C) treatment; SL (0 h to 48 h), leaves exposed to high salt (200 mM NaCl) treatment; SR (0 h to 72 h), roots exposed to high
salt (200 mM NaCl) treatment.
|
|
The expression patterns of AhGPATs in peanut leaves and roots after treatment with 200 mM NaCl were also monitored (Figure 6). The expression patterns of AhGPAT1, AhGPAT9 and AhATS1 were different in leaves and roots. Transcript levels of AhGPAT1 decreased distinctly and rapidly from 1 h to 48 h in the leaves of seedlings subjected to salt treatment, but increased obviously
in roots after 12 h treatment, with a nearly 4-fold increase 48 h after the salt treatment. The levels of AhGPAT9 transcript decreased gradually in leaves under salt stress, but increased obviously after 24 h treatment and exhibited nearly
a 7-fold increase after the roots were treated for 48 h. The transcript levels of AhATS1 decreased distinctly and rapidly from 1 h to 48 h in salt-treated leaves, but increased obviously in roots after 1 h treatment,
with a peak level of about 20-fold observed at 3 h. The expressions of AhGPAT2 were increased under salt stress, with a peak level at 3 h in both leaves and roots, where the greatest increases were about
3-fold and 41-fold, respectively. In leaves, the expressions of AhGPAT6 and AhGPAT8 decreased rapidly after 1h treatment and then increased to the peak level at 3 h. After 3 h, the levels of AhGPAT6 and AhGPAT8 transcripts decreased distinctly. In the roots, the expressions of AhGPAT6 and AhGPAT8 gradually increased under salt stress, with a maximum increase of about 4-fold observed at 3 h, and then decreased substantially.
A 20% solution of PEG-6000 was used to mimic drought stress to monitor the expression patterns of AhGPATs in peanut leaves and roots (Figure 7). In the leaves, the expressions of AhGPAT1 slightly increased 3 h after treatment, and then decreased from 6 h to 72 h. In PEG-treated roots, the levels of AhGPAT1 transcript were distinctly enhanced relative to the peak level (an approximately 9-fold increase) which was observed at 12
h. The transcript levels of AhGPAT2 were obviously increased in both leaves and roots under drought stress, with peak expression levels at 1 h in leaves and
24 h in roots. The greatest increase was about 16-fold in leaves and 5,537-fold in roots. Within 6 h after treatment, AhGPAT6 and AhGPAT8 genes were slightly down-regulated in leaves and obviously up-regulated in the roots of peanut seedlings subjected to drought
stress. The expressions of AhGPAT9 in leaves increased rapidly with a peak level of about 6-fold increase at 3 h under drought treatment, whereas in roots the
expressions increased slightly after 6 h treatment and then decreased from 6 h to 72 h. The expressions of AhATS1 were obviously increased in both leaves and roots under drought stress, with peak levels for both at 3 h. The greatest increase
was about 3-fold in leaves and 12-fold in roots.
|
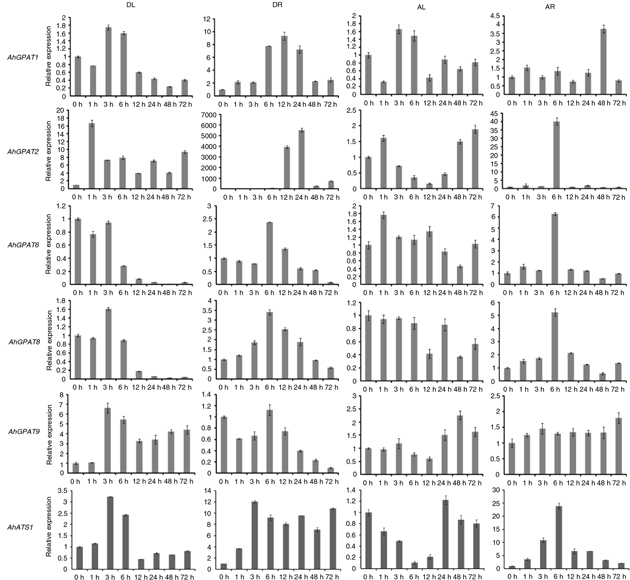 Figure 7. Expression analysis of six AhGPAT genes using qRT-PCR under drought and ABA stress, The relative mRNA abundance was normalized with respect to the peanut AhTUA5 gene. The bars were standard deviations (SD) of three technical repetitions. DL (0 h to 72 h), leaves exposed to 20% PEG-6000
treatment; DR (0 h to 72 h), roots exposed to 20% PEG-6000 treatment, AL (0 h to 72 h), leaves exposed to 100 uM ABA treatment;
AR (0 h to 72 h), roots exposed to 100 uM ABA treatment. Figure 7. Expression analysis of six AhGPAT genes using qRT-PCR under drought and ABA stress, The relative mRNA abundance was normalized with respect to the peanut AhTUA5 gene. The bars were standard deviations (SD) of three technical repetitions. DL (0 h to 72 h), leaves exposed to 20% PEG-6000
treatment; DR (0 h to 72 h), roots exposed to 20% PEG-6000 treatment, AL (0 h to 72 h), leaves exposed to 100 uM ABA treatment;
AR (0 h to 72 h), roots exposed to 100 uM ABA treatment.
|
|
We also examined the response of AhGPAT genes to exogenously applied ABA, which is a plant signaling molecule involved in plant defense signaling pathways (Figure 7). There was no obvious change in the levels of AhGPAT1 transcript in peanut leaves following ABA treatment, although the levels of AhGPAT1 transcript in roots were obviously higher 48 h after initial exposure to exogenous ABA. In leaves, the expressions of AhGPAT2 increased slightly after 1 h treatment with ABA and then decreased from 3 to 12 h. After 24 h, the levels of AhGPAT2 transcript remained slightly higher than in untreated leaves. The Levels of AhGPAT2 transcript were higher in ABA-treated roots than in untreated roots observed 6 h after treatment, with a maximum increase
of approximately 40-fold. There were no obvious changes in the abundances of AhGPAT6, AhGPAT8 and AhATS1 transcripts in peanut leaves after ABA treatment. However, the levels of three transcripts increased in roots, where they
reached maximum levels 6 h after ABA treatment, with the greatest increases observed being about 6-, 5- and 25-fold, respectively.
The expressions of AhGPAT9 were slightly increased in both the leaves and roots of seedlings subjected to ABA stress, with peak levels at 48 and 72
h, respectively.
The above results indicate that GPAT transcripts from peanuts are differentially expressed following exposure to abiotic stresses or abscisic acid. The levels
of AhGPAT2 transcript were distinctly enhanced after exposure to all four kinds of stress treatments except for ABA-treated leaves.
The transcripts of AhGPAT1, AhGPAT6, AhGPAT8 and AhATS1 increased substantially in roots exposed to salt, drought, and ABA stress. The expressions of AhGPAT6, AhGPAT8, AhGPAT9 and AhATS1 were slightly higher in leaves under certain stress conditions than under normal conditions. These results suggest that these
genes may play an important role in enhancing peanut resistance to abiotic stress. Some genes were obviously down-regulated
after stress treatments, such as AhGPAT1 and AhGPAT9 transcripts in cold- and salt-stressed leaves. This indicates that these genes may have a negative function in peanut abiotic
stress regulation.
4. DISCUSSIONTOP
sn-Glycerol-3-phosphate acyltransferase (GPAT) is an important enzyme in glycerolipid synthesis, and is involved in different
metabolic pathways and physiological functions. In this study, six genes were identified. These genes likely represent the
peanut homologues of Arabidopsis genes involved in the synthesis of cutin, suberin, membrane lipids, or storage lipids. Phylogenetic
analysis showed that AhATS1 fell into the plastidial ATS1 subgroup and showed a high sequence similarity with AtATS1. AhGPAT1 and AhGPAT2 belonged to the GPAT1–3 subfamily and shared high sequence similarities with AtGPAT1 and AtGPAT2/3, respectively. Sequence analysis indicates that the NH2-terminal domain of the three genes contains four acyltransferase motifs (Pfam 01553) that are conserved among glycerolipid
acyltransferase family members, which include GPATs, AGPATs, and a dihydroxyacetone-phosphate acyltransferase (Takeuchi and
Reue, 2009). It has been suggested that motifs I and IV are important for catalysis, and that motifs II and III are important for substrate
binding. The COOH-terminal domain is also necessary for enzyme activity and appears to physically interact with the NH2-terminal domain to contribute to either catalysis or substrate binding (Pellon-Maison et al., 2006).
The AhGPAT6 and AhGPAT8 proteins belonged to the GPAT4/6/8 clades and shared high sequence similarity with AtGPAT4/8 and AtGPAT6, respectively. Sequence analysis reveals that AhGPAT6 and AhGPAT8 each contain an N-terminal HAD-like domain attached to the acyltransferase moiety. The HAD domain is widespread over the
three super-kingdoms and is found in a very diverse range of enzymes with hydrolytic activities. Maximum homology of the HAD
domain from GPATs out of plants is seen for members of the “PSP/P5N-1 assemblage” (Burroughs et al., 2006), which are characterized by the presence of a C1-type cap module with a four-helix arrangement. This group includes enzymes
with activities as diverse as those of phosphoserine phosphatases (PSP family) and nucleotidases (P5N-1 family). The presence
of this typical hydrolytic domain in plant GPATs allows them to behave as bifunctional enzymes that catalyze the dephosphorylation
of glycerol in addition to acyl transfer, thus yielding MAGs as the reaction product (Mañas-Fernández et al., 2010).
The AhGPAT9 protein showed high sequence similarity to AtGPAT9, which was identified in Arabidopsis by a bioinformatics approach, and exhibits a much closer evolutionary relationship with
mammalian GPATs. Although the enzymatic activity of AtGPAT9 has not been directly confirmed and its physiological function is unknown, polypeptide sequence alignment, phylogenetic analysis,
conserved domain analysis and gene expression data have all suggested that AtGPAT9 may play an essential role in the synthesis
of membrane and storage lipids in plants (Gidda et al., 2009; Chen et al., 2011b). Expression profiling revealed that the levels and tissue-specific accumulations of AhGPAT9 transcript are distinct from those of other GPAT family members, which is consistent with the more diverged nature of the AtGPAT9 gene. Notably, the expression patterns of AhGPAT9 coincided with the lipid accumulation rate in peanut seed. This suggests a potential role for AhGPAT9 in glycerolipid metabolism in developing seeds, although this possibility remains to be tested experimentally.
Cutin and suberin are extracellular lipid barriers deposited by certain types of plant cells (Yang et al., 2012). They are both fatty acid–and glycerol-based extracellular polymers that are insoluble in water and organic solvents (Beisson et al., 2007). These insoluble polymers and other associated waxes function to control water, gas, and ion fluxes and serve as physical
barriers to protect plants from pathogen invasion (Schreiber, 2010). The seed coats of Arabidopsis gpat5 mutants were substantially more permeable to tetrazolium salts than those of wild-type seeds. Furthermore, the germination
rate of gpat5 seeds under high salt was reduced, and gpat5 seedlings were less tolerant of salt stress than wild-type seedlings (Beisson et al., 2007). The lines of B. napus in which GPAT4 expression was suppressed using RNAi exhibited alterations in cuticle load and stomatal structure, resulting in increased
water loss (Chen et al., 2011b). Our results indicated that AhGPAT2 was distinctly enhanced under all four kinds of stress treatments except for ABA-treated leaves. The levels of AhGPAT1 transcript and cutin-associated AhGPAT6 and AhGPAT8 transcripts increased substantially in the roots of seedlings subjected to salt, drought, and ABA stresses. Thus, we infer
that these GPAT genes may be involved in regulating some kinds of abiotic stress in peanuts.
GPAT family proteins play crucial roles in the synthesis of cutin, suberin, membrane lipids, and storage lipids (Chen et al., 2011a). Better the understanding of this enzyme family will be valuable to efforts to modify the content and composition of seed
oils or to improve abiotic stress resistance in plants. The information generated in our study has improved our understanding
of the involvement of these genes in lipid synthesis and opens the way to selecting candidate genes for functional validation
studies in peanuts.
ACKNOWLEDGMENTSTOP
This study was supported by grants from the China Agriculture Research System (CARS-14), the National Natural Science Foundation
of China (31000728; 31100205; 31200211), the Natural Science Fund of Shangdong Province (ZR2009DQ004; ZR2011CQ036; ZR2014YL012),
the Promotive Research Fund for Young and Middle-aged Scientisits of Shandong Province (BS2010NY023), Qingdao Municipal Science
and Technology Plan Project (11-2-4-9-(3)-jch; 11-2-3-26-nsh; 12-1-4-11-(2)-jch), the Fund of the Key Laboratory of Biology
and Genetic Improvement of Oil Crops, Ministry of Agriculture (2014010).
REFERENCESTOP
| ○ |
Ariizumi T, Kishitani S, Inatsugi R. 2002. An increase in unsaturation of fatty acids in phosphatidylglycerol from leaves
improves the rates of photosynthesis and growth at low temperatures in transgenic rice seedlings. Plant. Cell. Physiol. 43, 751–758. http://dx.doi.org/10.1093/pcp/pcf087.
|
| ○ |
Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. 2007. The acyltransferase GPAT5 is required for the synthesis of suberin
in seed coat and root of Arabidopsis. Plant. Cell. 19, 351–368. http://dx.doi.org/10.1105/tpc.106.048033.
|
| ○ |
Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. 2006. Evolutionary genomics of the HAD superfamily: understanding the
structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 361, 1003–1034. http://dx.doi.org/10.1016/j.jmb.2006.06.049. |
| ○ |
Cao J, Li JL, Li D, Tobin JF, Gimeno RE. 2006. Molecular identification of microsomal acyl-CoA: glycerol-3-phosphate acyltransferase,
a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 103, 19695–19700. http://dx.doi.org/10.1073/pnas.0609140103.
|
| ○ |
Chen X, Snyder CL, Truksa M, Shah S, Weselake RJ. 2011a. sn-Glycerol-3-phosphate acyltransferases in plants. Plant Signaling Behavior. 6, 1695–1699. http://dx.doi.org/10.4161/psb.6.11.17777.
|
| ○ |
Chen X, Truksa M, Snyder CL, El-Mezawy A, Shah S, Weselake RJ. 2011b. Three Homologous genes encoding sn-glycerol-3-phosphate
acyltransferase 4 exhibit different expression patterns and functional divergence in Brassica napus. Plant Physiol. 155, 851–865. http://dx.doi.org/10.1104/pp.110.169482.
|
| ○ |
Chi XY, Hu RB, Yang QL, Zhang XW, Pan LJ, Chen N, Chen MN, Yang Z, Wang T, He YN, Yu SL. 2012. Validation of reference genes
for gene expression studies in peanut by quantitative real-time RT-PCR. Mol. Genet. Genomics. 287, 167–176.
|
| ○ |
Chi XY, Yang QL, Pan LJ, Chen MN, He YN, Yang Z, Yu SL. 2011. Isolation and characterization of fatty acid desaturase genes
from peanut (Arachis hypogaea L.). Plant. Cell. Rep. 30, 1393–1404. http://dx.doi.org/10.1007/s00438-011-0665-5.
|
| ○ |
Gidda SK, Shockey JM, Rothstein SJ, Dyer JM, Mullen RT. 2009. Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine
ER retrieval motif in plant cells. Plant. Physiol. Biochem. 47, 867–879. http://dx.doi.org/10.1016/j.plaphy.2009.05.008.
|
| ○ |
Gonzalez-Baró MR, Lewin TM, Coleman RA. 2007. Regulation of Triglyceride Metabolism II. Function of mitochondrial GPAT1 in
the regulation of triacylglycerol biosynthesis and insulin action. Am. J. Physiol. Gastrointest. Liver. Physiol. 292, G1195–G1199. http://dx.doi.org/10.1152/ajpgi.00553.2006.
|
| ○ |
Harwood H. 1984. Oleochemicals as a fuel. Mechanical and economic feasibility. J. Am. Oil Chem. Soc. 61, 315–324. http://dx.doi.org/10.1007/BF02678788.
|
| ○ |
Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta
Delta C(T)) Method. Methods. 25, 402–408. http://dx.doi.org/10.1006/meth.2001.1262.
|
| ○ |
Li XC, Zhu J, Yang J, Zhang GR, Xing WF, Zhang S, Yang ZN. 2011. Glycerol-3-phosphate acyltransferase 6 (GPAT6) is important
for tapetum development in Arabidopsis and plays multiple roles in plant fertility. Molecular Plant. 5, 131–142. http://dx.doi.org/10.1093/mp/ssr057.
|
| ○ |
Li Y, Beisson F, Koo AJ, Molina I, Pollard M, Ohlrogge J. 2007b. Identification of acyltransferases required for cutin biosynthesis
and production of cutin with suberin-like monomers. Proc. Natl. Acad. Sci. USA. 104, 18339–18344. http://dx.doi.org/10.1073/pnas.0706984104.
|
| ○ |
Li Y, Beisson F, Ohlrogge J, Pollard M. 2007a. Monoacylglycerols are components of root waxes and can be produced in the aerial
cuticle by ectopic expression of a suberin-associated acyltransferase. Plant. Physiol. 144, 1267–1277. http://dx.doi.org/10.1104/pp.107.099432.
|
| ○ |
Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F. 2009. Nanoridges that characterize the surface morphology
of flowers require the synthesis of cutin polyester. Proc. Natl. Acad. Sci. USA. 106, 22008–22013. http://dx.doi.org/10.1073/pnas.0909090106.
|
| ○ |
Mañas-Fernández A, Li-Beisson Y, Alonso DL, García-Maroto F. 2010. Cloning and molecular characterization of a glycerol-3-phosphate
O-acyltransferase (GPAT) gene from Echium (Boraginaceae) involved in the biosynthesis of cutin polyesters. Planta. 232, 987–997. http://dx.doi.org/10.1007/s00425-010-1232-8.
|
| ○ |
Murata N, Ishizaki-Nishizawa O, Higashi S, Hayashi H, Tasaka Y, Nishida I. 1992. Genetically engineered alteration in the
chilling sensitivity of plants. Nature. 365, 710–713. http://dx.doi.org/10.1038/356710a0.
|
| ○ |
Murata N, Tasaka Y. 1997. Glycerol-3-phosphate acyltransferase in plants. Biochim. Biophys. Acta. 1348 (1–2), 10–16. http://dx.doi.org/10.1016/S0005-2760(97)00115-X.
|
| ○ |
Nishida I, Tasaka Y, Shiraishi H, Murata N. 1993. The gene and the RNA for the precursor to the plastid-located glycerol-3-phosphate
acyltransferase of Arabidopsis thaliana. Plant. Mol. Biol. 21, 267–277. http://dx.doi.org/10.1007/BF00019943.
|
| ○ |
Pellon-Maison M, Coleman RA, Gonzalez-Baró MR. 2006. The C-terminal region of mitochondrial glycerol-3-phosphate acyltransferase-1
interacts with the active site region and is required for activity. Arch. Biochem. Biophys. 450, 157–166. http://dx.doi.org/10.1016/j.abb.2006.03.009.
|
| ○ |
Schreiber L. 2010. Transport barriers made of cutin, suberin and associated waxes. Trends. Plant. Sci. 15, 546–553. http://dx.doi.org/10.1016/j.tplants.2010.06.004.
|
| ○ |
Sui N, Li M, Zhao SJ, Li F, Liang H, Meng QW. 2007. Overexpression of glycerol-3-phosphate acyltransferase gene improves chilling
tolerance in tomato. Planta. 226, 1097–1108. http://dx.doi.org/10.1007/s00425-007-0554-7.
|
| ○ |
Takeuchi K, Reue K. 2009. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis.
Am. J. Physiol. Endocrinol. Metab. 296, E1195–E1209. http://dx.doi.org/10.1152/ajpendo.90958.2008.
|
| ○ |
Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 24, 1596–1599. http://dx.doi.org/10.1093/molbev/msm092.
|
| ○ |
Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through
sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids. Res. 22, 4673–4680. http://dx.doi.org/10.1093/nar/22.22.4673.
|
| ○ |
Xu C, Cornish AJ, Froehlich JE, Benning C. 2006. Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants
deficient in acyl-ACP glycerol-3-phosphate acyltransferase. Plant. J. 47, 296–309. http://dx.doi.org/10.1111/j.1365-313X.2006.02790.x.
|
| ○ |
Yan K, Chen N, Qu YY, Dong XC, Meng QW, Zhao SJ. 2008. Overexpression of sweet pepper glycerol-3-phosphate acyltransferase gene enhanced thermotolerance of photo-synthetic apparatus in transgenic tobacco. J. Integrative Plant Biol. 50 613–621. http://dx.doi.org/10.1111/j.1744-7909.2008.00652.x.
|
| ○ |
Yang WL, Simpson JP, Li-Beisson YH, Beisson F, Pollard M, Ohlrogge JB. 2012. A land-plant-specific glycerol-3-phosphate acyltransferase
family in Arabidopsis: substrate specificity, sn-2 preference and evolution. Plant Physiol. 160, 638–652. http://dx.doi.org/10.1104/pp.112.201996.
|
| ○ |
Zheng Z, Xia Q, Dauk M, Shen W, Selvaraj G, Zou J. 2003. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation
and male fertility. Plant Cell. 15, 1872–1887. http://dx.doi.org/10.1105/tpc.012427.
|
Figure 1. Amino acid alignment of peanut plastidial ATS1 proteins and closely related proteins found in the GenBank. Residues shared
by a fraction of sequences above 0.5 were shaded, identical residues in black, similar residues in grey. AT-like domains were
boxed (ATI to AT-IV). Critical residues previously identified in similar proteins were marked by dots (binding site in AT
domain) or triangles (catalytic residues in AT domain). GenBank accession numbers were as follows: Arachis hypogaea (AhATS1, KC762933), Arabidopsis thaliana (AtATS1, NP_174499), Glycine max (GmATS1, XP_003516958), Physcomitrella patens (PpATS1, XP_001771299), Medicago truncatula (MeATS1, XP_003612801), Chlamydomonas reinhardtii (CrATS1, XP_001694977), Volvox carteri (VoATS1, XP_002950506).