Authentication of olive oil based on DNA analysis
A. Batrinoua,*, I.F. Stratia,*, D. Houhoulaa, J. Tsaknisa and V.J. Sinanogloua
aDepartment of Food Science and Technology Faculty of Food Sciences University of West Attica, Athens, Greece
*Corresponding authors: batrinou@uniwa.gr; estrati@uniwa.gr
| |
SUMMARY
Olive oil, which has been produced mainly in the Mediterranean area since the ancient times, has a high nutritional value linked to many health benefits. Extra virgin, which is the purest form of olive oil, has excellent quality and premium prices. Many cases of adulteration and fraud necessitate the development of reliable and accurate methods for olive oil authentication. DNA-based methods analyze the residual DNA extracted from olive oil and use molecular markers for genetic identification of different species, subspecies or cultivars because these markers act as signs which reflect distinct genetic profiles. This study reviews the process by which DNA from olive oil is extracted and analyzed by the most recently used markers in the authentication of olive oil, such as Simple Sequence Repeats (SSR) or microsatellites and the single nucleotide polymorphisms (SNPs). Methods of analysis such as qPCR and digital PCR are also discussed with a special emphasis placed on the method of High-Resolution Melting (HRM), a post-polymerase chain reaction method, which enables rapid, high performing identification of genetic variants in the DNA regions of interest without sequencing, and may differentiate very similar cultivars which differ in only one nucleotide in a specific locus.
|
| |
RESUMEN
Autenticación de aceite de oliva mediante análisis de ADN. El aceite de oliva, producido principalmente en el área mediterránea desde la antigüedad, tiene un alto valor nutricional vinculado a muchos beneficios para la salud. El aceite de oliva virgen extra, que es la forma más pura de aceite de oliva, tiene una excelente calidad y precios premium. Muchos casos de adulteraciones y fraudes requieren el desarrollo de métodos fiables y precisos para la autenticación del aceite de oliva. Los métodos basados en el ADN analizan el ADN residual extraído del aceite de oliva y usan marcadores moleculares para la identificación genética de diferentes especies, subespecies o cultivares, porque estos marcadores actúan como signos que producen perfiles genéticos distintos. Este estudio revisa el proceso mediante el cual el ADN del aceite de oliva es extraído y analizado por los marcadores utilizados más recientemente en la autenticación del aceite de oliva, como las repeticiones de secuencia simple (SSR) o los micro satélites y los polimorfismos de un solo nucleótido (SNP). Los métodos de análisis como qPCR y PCR digital también se analizan haciendo especial énfasis en el método de fusión de alta resolución (HRM), un método de reacción en cadena posterior a la polimerasa, que permite la identificación rápida y con alto rendimiento de variantes genéticas en regiones del ADN de interés sin secuenciación, y pueden diferenciar cultivares muy similares, que difieren en un solo nucleótido, en un lugar específico.
|
1. INTRODUCTIONTOP
The cultivation of olive trees began thousand years ago in the Mediterranean area (fossil pollen was found in Macedonia, Greece and fossilized leaves were found on the island of Santorini, Greece, dated to 37,000 BP (NCBI)) and it has developed into the most important oil-producing crop in the Mediterranean region, covering more than 10 million hectares of land worldwide (FAOSTAT, 2016). The high nutritional value and health benefits of olive oil and in particular Virgin olive oil (VOO) represent a hallmark of the Mediterranean Diet, and have been established by various studies (Parkinson and Cicerale, 2016; Martinez-Gonzalez and Martin-Calvo, 2016; Trichopoulou et al., 2014). The European Commission has recently permitted health claims concerning olive oil polyphenols, especially hydroxytyrosol and its derivatives (e.g. oleuropein complex and tyrosol) (Regulation EU 432/2012).
The growing demand for olive oil and the high prices consumers are willing to pay for high quality products such as extra virgin olive oil (EVOO) have resulted in many cases of adulteration and fraud in recent years (Bajoub et al., 2018; Ramos-Gomez et al., 2016). The European Union has applied laws regulating the classification and labeling of olive oil in order to protect its geographical origin and quality. Commercial olive oils that are produced from a specific region and follow a particular traditional production process may be labeled as Protected Designation of Origin (PDO) or Protected Geographical Indication (PGI) (European Council Regulation, 2006).
The olive tree belongs to the species Olea europaea, which is actually a complex (the olive complex) comprising of six subspecies and more than 1250 cultivars that have different genotypes (differences in the sequence of DNA) (Bracci et al., 2011). The Olea europaea subsp. europaea, which has spread across the Mediterranean basin, has 46 chromosomes and is diploid (has two copies of each chromosome) (Besnard et al., 2008; Besnard et al., 2009). The high number of different olive genotypes which are currently cultivated necessitates the application of DNA-based methods that use genetic markers to assess varietal identification, genetic authenticity, traceability and adulteration (Sebastiani and Busconi, 2017).
DNA-based methods that test the authenticity of foods rely on the fact that the residual DNA extracted from any food matrix can be analyzed and provides information about the genetic identity of any species, subspecies or variety present in the matrix. In the case of olive oil, DNA analysis can reveal the presence of unique DNA sequences of different plant species (e.g. hazelnut oil in olive oil, Vietina et al., 2013), different varieties (e.g. the presence of non-PDO cultivars in a product claiming to be PDO monovarietal, Pasqualone et al., 2007) and can also resolve taxonomic issues such as synonymies (different names for the same variety), homonymies (same name for different variety) and misnaming (Muzzalupo et al., 2015), thus providing a powerful tool for discrimination analysis (Sebastiani and Busconi, 2017; Kalogianni et al., 2015; Madesis et al., 2014; Galimberti et al., 2013; Ben-Ayed et al., 2013).
The unique DNA sequences found in each organism (and thus in each plant), which are called genetic or molecular markers, are specific sequences of DNA with known physical locations on chromosomes and they constitute points of variation that can be used to identify individuals or species (NCI 2019, Nature 2019).
The common genetic markers which have been used for the identification of olive cultivars and the traceability of olive oil are the “simple sequence repeats” (called SSR or microsatellites, which are tandem repeats of di-, tri-, tetra-, and penta-nucleotides, Bazakos et al., 2016a; Kalogianni et al., 2015) and the “single nucleotide polymorphisms” (called SNPs, Abuzayed et al., 2018; Mousavi et al., 2017). Other markers have also been used as techniques to identify genetic variation, such as “randomly amplified polymorphic DNA” (called RAPD consisting of 10-mer primers that randomly anneal to several loci leading to the amplification of many sequences by PCR, providing many markers per assay, Belaj et al., 2003, Muzzalupo and Perri, 2002), “amplified fragment length polymorphism” (called AFLP which is based on the detection of restriction fragments by PCR amplification, Montemurro et al., 2008; Pafundo et al., 2005) and “inter-simple sequence repeat” (called ISSR, which are DNA fragments of about 100–3,000 bp present at an amplifiable distance in between two identical microsatellite repeat regions oriented in the opposite direction, Terzopoulos et al., 2005; Gemas et al., 2004; Pasqualone et al., 2001). RAPD, AFLP and ISSR have advantages when analyzing genetic relationships in olive trees mainly because the design of primers for the PCR do not require prior knowledge of the genome sequence. However, these markers have the drawback of low reproducibility in olive oil studies (reviewed by Pasqualone et al., 2016; Sebastiani and Busconi, 2017; Bazakos et al., 2016, Ben-Ayed et al., 2013). The genetic markers used by the most recent studies for olive oil authentication are simple sequence repeats (SSR) and single nucleotide polymorphisms (SNPs) (Pasqualone et al., 2016) and are located either in nuclear DNA or in the DNA found in organelles (chloroplast or mitochondrial DNA) (Pereira et al., 2018).
Once the DNA has been efficiently recovered from a sample, a specific region of DNA (a target sequence which is usually a genetic marker) is amplified in many copies using the Polymerase Chain Reaction (PCR) so that an adequate amount of DNA is formed to permit further analysis. The next step is generally called “genotyping” and involves a number of methods by which differences in the DNA sequence of the sample are visualized and identified (Figure 1). Differences in the sequence of DNA (called variants or polymorphisms, which are any difference in the nucleotide sequence between individuals) can be single-base pair changes (nucleotide substitutions), deletions, insertions, or even variations in the number of copies of a given DNA sequence (Karki et al., 2015).
|
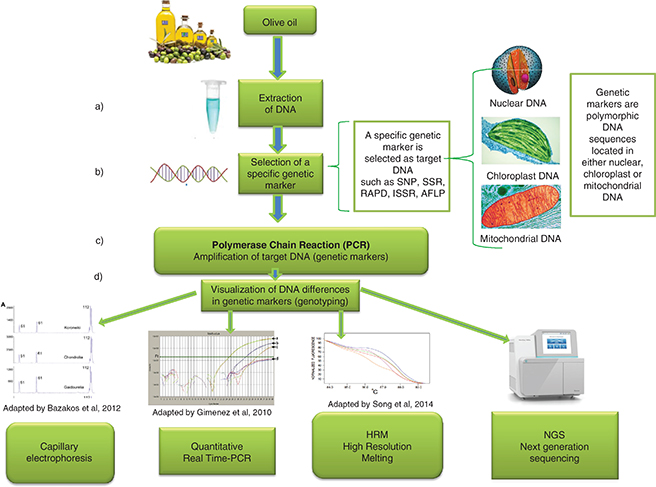 Figure 1. Simplified diagram of DNA analysis of olive oil: a) DNA extraction of the olive oil, b) selection of specific genetic markers as target DNA which will enable discrimination among cultivars, establish genetic authenticity, or detect adulteration with different species. These genetic markers are polymorphic DNA sequences located in either nuclear, chloroplast or mitochondrial DNA and could be SNPs, SSRs, RAPD, ISSR, AFLP, etc. c) application of PCR with the appropriate primers which amplify the target DNA, d) genotyping or visualization of amplified target sequences by methods that can distinguish differences, even in one base, among DNA sequences. Figure 1. Simplified diagram of DNA analysis of olive oil: a) DNA extraction of the olive oil, b) selection of specific genetic markers as target DNA which will enable discrimination among cultivars, establish genetic authenticity, or detect adulteration with different species. These genetic markers are polymorphic DNA sequences located in either nuclear, chloroplast or mitochondrial DNA and could be SNPs, SSRs, RAPD, ISSR, AFLP, etc. c) application of PCR with the appropriate primers which amplify the target DNA, d) genotyping or visualization of amplified target sequences by methods that can distinguish differences, even in one base, among DNA sequences.
|
|
For genotyping (identifying the base sequence and thus revealing the polymorphisms), PCR amplicons (copies) of target-DNA can be sequenced, or analyzed by agarose gel electrophoresis or high-resolution capillary electrophoresis (CE). Capillary electrophoresis has a much lower limit of detection for DNA fragments. It can discriminate DNA sequences which differ by only few nucleotides in length and is more automated compared to agarose gel electrophoresis (Alvarez-Rivera et al., 2018; Lian and Zeng, 2017; Spaniolas et al., 2010; 2008). Techniques such as quantitative real-time PCR or high-resolution melting analysis (HRM) are increasingly used as they offer the advantage of simultaneous PCR amplification and genotyping, as well as the technique of Next-Generation Sequencing (NGS), which provides massive data of parallel short-read per run, lowering the analysis cost and revealing DNA fragments from a variety of species at the same time (Lo and Shaw, 2018; Madesis et al., 2014).
This review analyses the latest advances in the application of DNA-based methods in the authentication of olive oil with a special emphasis on the use of the more recently developed species-specific molecular markers such as simple sequence repeats (SSR) and single nucleotide polymorphisms (SNPs) and high-throughput methods such as High-Resolution Melting (HRM) as methods that present numerous advantages for varietal identification.
2. EXTRACTION OF DNA TOP
Food genomics are commonly applied for assessing food safety, authenticity and traceability due to the many advantages of the DNA-based methodologies compared to other techniques based on the analysis of metabolites. The first step in all DNA-based authenticity tests is the extraction of DNA from the sample to be analyzed. DNA is a thermally stable molecule and can be detected even in small fragments in any type of raw, cooked or processed food; whereas proteins are denatured under high temperatures or when processed.
In olive oil, the main advantage to extracting DNA is that its sequence is not influenced by environmental factors; whereas the metabolic profile of olive oil such as polyphenol content and fatty acids is strongly affected by environmental factors (e.g. climate conditions and soil composition) as well as other factors such as stage of ripeness, harvesting time, storage period and extraction method (Ramos-Gómez et al., 2016; Kalaitzis and El-Zein, 2016; Ben-Ayed et al., 2013; Agrimonti et al., 2011; Montealegre et al., 2010; Heimler et al., 2004). Therefore, the base sequence of DNA extracted from olive oil can be used as a genetic fingerprint to reveal the identities of different oils that might be present in the oil. However, one of the major limiting steps of the process is that DNA extracted from olive oil can be very fragmented and may contain phenolic compounds or residual polysaccharides which can act as inhibitors of DNA polymerase which may affect the quality of the PCR reaction by yielding non-specific PCR amplifications (Raieta et al., 2015). However, recent studies have reported the development of a DNA polymerase enzyme which is more tolerant to PCR inhibitors and can be applied directly to crude tissues (Chiappetta et al., 2017; Muzzalupo et al., 2015). Another limitation may be the discrepancy between the results obtained from the DNA extracted from leaves as opposed to
the DNA extracted from oils, which suggests differences in the quality of the extracted DNA (Ramos-Gómez et al., 2016; Pasqualone et al., 2007; Pasqualone et al., 2016). The correspondence between the profiles of microsatellites of oil and leaves can be significantly improved by shortening the target sequences for PCR amplification to as low as 80 bp (Ramos-Gomez et al., 2014; Ramos-Gómez et al., 2016; Bracci et al., 2011). The amount and quality of DNA isolated from oil samples remain critical parameters for the success of subsequent analyses (Scollo et al., 2016). DNA extraction from olive oil has been successfully achieved by various protocols such as CTAB and commercial kits such as Wizard kit (Promega), QIAamp DNA stool extraction kit (Qiagen), Nucleospin Food and Nucleospin Plant (Macherey Nagel), as reviewed by Ben-Ayed et al., (2013) and Agrimonti et al., (2011).
3. SIMPLE SEQUENCE REPEATS (SSR)TOP
Simple Sequence Repeats (SSR) or microsatellites are DNA sequences of 1 to 10 nucleotides long. They occur as tandem repeats (TRs) of di-, tri-, tetra-, and penta-nucleotides and are ubiquitous in eukaryotic genomes (Gupta et al., 1994). SSRs are widely distributed throughout the genome, especially in the euchromatin of eukaryotes, and in the coding and non-coding DNA of the nucleus and organelles. SSRs have been extensively used as markers for genotyping plants and olive oil varieties over the past 15 years (Table 1) because they are codominant (the alleles in a gene pair in a heterozygote are both expressed), multi-allele (multiple alleles are found in the population of the same species) and highly informative genetic markers which are experimentally reproducible and commonly found among related species (Ben-Ayed et al., 2013; Ben-Ayed et al., 2016). The occurrence of SSRs is usually lower in gene regions, probably due to the fact that SSRs have a high mutation rate that could compromise gene expression. SSRs exhibit repeat polymorphisms that usually result from the addition or deletion of the entire repeat units or motifs. Therefore, different individuals exhibit variations (polymorphisms) in SSRs as differences in the repeat numbers of specific DNA sequences (Vieira et al., 2016). An example in Olea europaea is the microsatellite (GA)n, which is the dinucleotide GA repeated n times in a specific region of a specific chromosome (genetic locus). The polymorphism lies in the fact that different varieties of the same species may have a different number of repeats, e.g. one SSR locus is named ssrOeUA-DCA1 (Sefc et al., 2000) and has 22 repetitions of the dinucleotide GA symbolized as (GA)22 but this allele is polymorphic, meaning that in some individuals (olive trees) a different number of repetitions is found on the same locus. Therefore, by identifying the number of repeats found in the DNA isolated from a sample, a process performed by amplifying the target sequence by PCR amplification using specific primers, discrimination of the variety can be achieved. Usually, a combination of SSR markers is used to provide a more powerful discriminatory analysis (Bazakos et al., 2016). Some applications include expressed sequence tag SSR (EST-SSR), which derive from regions of the genome that are expressed (sequence coding for a specific protein) and therefore these regions have greater transferability among species and their variation can be associated with the phenotype (Duran et al., 2009).
Table 1. Applications of SSR (Simple Sequence Repeat) in the authentication of olive oil
| Year of publication |
Main Findings |
Reference |
| 2018 |
SSR markers were used to assess the genetic diversity and population structure of 19 olive cultivars (15 from Palestine and 4 from Turkey).
|
Abuzayed et al., 2018 |
| 2017 |
The genotypes of 370 olive trees were characterized into 59 distinct genotypes by using 10 standard dinucleotide SSR markers and nine new EST-SSR markers. |
Mousavi et al., 2017 |
| 2015 |
PCR was used to generate SSR and ISSR markers from genomic DNA extracted from young leaves and a total of 107 polymorphic bands were detected on thirteen SSR loci, with an average of eight alleles per locus. |
Zhan et al., 2015 |
| 2015 |
SSR markers discriminated olive cultivars efficiently and clustering results based on 20 SSR markers and nine consensus SSR primers were highly correlated. No concordance was found between clustering of olive cultivars based on SSR markers and fatty acid composition of olive oil. |
Ipek et al., 2015 |
| 2014 |
33 SSR markers and 11 morphological characteristics of the endocarp were used to characterize and authenticate 824 trees, representing 499 accessions from 21 countries of origin, from the World Olive Germplasm Bank of the Cordoba collection. |
Trujillo et al., 2014 |
| 2014 |
Eight informative microsatellite markers were used for fingerprinting and evaluatinggenetic similarity among 15 Tunisian olive cultivars. |
Ben Ayed et al., 2014 |
| 2012 |
One microsatellite marker was adequate to discriminate Leucocarpa olive oil from six other monovarietal olive oils. |
Pasqualone et al., 2012 |
| 2011 |
Traceability of 21 mono-varietal olive oils using microsatellites. |
Vietina et al., 2011 |
| 2009 |
A single, highly polymorphic SSR marker could discriminate seven Italian PDO mono-varietal olive oils. |
Alba et al., 2009 |
| 2007 |
Verification of the identity of the Ogliarola salentina cultivar in the PDO Italian extra virgin olive oil (Collina di Brindisi) by the genotyping of a limited number of DNA microsatellites. |
Pasqualone et al., 2007 |
| 2007 |
DNA purified from oil was used for microsatellite analysis and the profiles of DNA extracted from a monovarietal oil and from the leaves of the same cultivar matched. |
Muzzalupo et al., 2007 |
| 2004 |
PCR with different SSR primer sets was used to detect alien alleles in commercial virgin oil samples of known composition, either single cultivars or mixtures of them. |
Breton et al., 2004 |
4. SINGLE NUCLEOTIDE POLYMORPHISMS (SNPS)TOP
Single nucleotide polymorphisms (SNPs) are the most common type of DNA polymorphism in most organisms and they have gained popularity as forensic markers in all areas of biomedical and genetic research. SNPs can discriminate samples of the same species or variety which differ in only one nucleotide in a specific genetic locus (a locus could be any coding or non-coding region in the DNA sequence of an organism). An example of a SNP would be if a cytosine (C) nucleotide is present at a particular locus in one individual’s DNA but a thymine (T) nucleotide occurs at the same locus in another individual’s DNA.
The advantages to SNPs also apply to olive oil authentication: a) they are the most abundant genetic markers; b) they have lower mutation rates and are stably inherited; c) they can be detected within a low size range of amplicons (e.g. 100bp) so they are a better choice for DNA that has been degraded and extracted from a complex matrix such as olive oil, d) they are mostly biallelic (a specific locus in a genome contains two observed alleles), e) they are able to differentiate very similar cultivars that may differ in only one nucleotide in a specific locus (Mehta et al., 2017; Kalogianni et al., 2015; Bracci et al., 2011; Agrimonti et al., 2011).
Most of the methods used to identify the points of variations, therefore genotyping SNPs, rely on amplification by the PCR of the genomic regions that span the SNPs before the actual genotyping reaction. Also, the PCR is sensitive and specific enough to distinguish between heterozygous and homozygous SNP genotypes in large, complex genomes (Syvanen, 2001).
5. QUANTITATIVE REAL-TIME PCR (QPCR) IN AUTHENTICATION OF OLIVE OILTOP
The quantification of an ingredient is essential in order to verify an adulteration, whether intended or unintended, or to evaluate if a limit set by law has been exceeded (Madesis et al., 2014). Quantitative Real-time PCR (qPCR) measures the number of copies of the marker gene (the gene that codes for the ingredient or adulterant) and compares it to the copy number of a ‘normalizing’ gene, thus providing a percentage of the ingredient or adulterant (Primrose et al., 2010). qPCR is more sensitive than conventional PCR and enables the detection and quantification of DNA in very low amounts. Another advantage is that the molecular markers used in qPCR are designed to amplify small fragments of DNA; therefore, the problems resulting from DNA degradation are reduced. Many studies have used quantitative Real-time PCR in the authentication of edible
oils (Ramos-Gómez et al., 2014; Agrimonti et al., 2011; Giménez et al., 2010). Oils which belong to the Protected Designation of Origin (PDO) and Protected Geographical Indication (PGI) do not usually consist of a single variety (monovarietal) so quantification is essential to determine the proportions of the components of the mixtures of varietal types. Ramos-Gomez et al., (2016) outlined an effective qPCR system for olive detection in oils and derived foods based on the relative quantification of olive DNA.
This system (named PetN–PsbM system after the PetN gene encoding a cytochrome b6/f complex subunit N and the PsbM gene encoding a photosystem II protein M) detected by amplification of DNA as low as 0.1% of olive content and olives as ingredient in 12 commercial food products. A qPCR method based on the chloroplast trnL (UAA) was used to establish a method for the authentication of olive oil (Alonso-Rebollo et al., 2017) and in another study the presence of an adulterant at a relative content as low as 5% was successfully determined with a chloroplast trnL (UAA) intron gene PCR-CE method; whereas the analysis of fatty acids failed to detect fraud when hazelnut oil was used as the adulterant (Uncu et al., 2017).
Droplet digital-PCR (ddPCR) is a more recent, promising technique that permits the absolute quantification of DNA and is considered more precise and reproducible, especially in samples with low levels of nucleic acids (Taylor et al., 2017). In this method, the PCR products are partitioned in monodispersed droplets which are read through a droplet flow cytometer and the absolute quantities of target DNA are measured (Hindson et al., 2011). Scollo et al., (2016) extracted DNA from olive oil with four different extraction protocols and successfully quantified olive DNA with ddPCR.
6. HIGH RESOLUTION MELTING (HRM)TOP
High-resolution melting (HRM) analysis is a post-polymerase chain reaction (PCR) method that enables rapid, high-throughput identification of genetic polymorphisms in the target-DNA without sequencing. In the first step, PCR is performed to amplify the target DNA (region of interest) in many copies in the presence of a fluorescent dye that binds to double-stranded DNA (dsDNA). This dye emits fluorescence at a high intensity when it is bound in the double-stranded DNA but when DNA is thermally denatured and is converted to single-stranded random coils the dye shows low levels of fluorescence (unbound state). The thermal denaturation step occurs after PCR is completed and the amplicon (usually a 50 to 500 bp long dsDNA sequence amplified in many copies) is denatured by gradually increasing temperature (ca. by 0.01 oC to 0.2 oC). During this step, termed «melting analysis» (melting meaning thermal denaturation of dsDNA), the gradually denaturing amplicon releases the fluorescent dye.
The melting temperature (Tm = the temperature at which 50% of the DNA is single- stranded) varies among DNA molecules and depends on the length of the DNA sequence and its Guanine and Cytosine (GC) base content (in the double-stranded DNA Guanine and Cytosine bases are bound with three hydrogen bonds and are more stable than Adenine and Thymine, which are bound with 2 hydrogen bonds).Therefore, DNA sequences which have a high GC content demonstrate a higher Tm than DNA sequences that contain a low number of GC base pairs. A curve, called melting curve, is obtained from plotting the decreasing fluorescent emission values against the increasing temperature (Figure 2). This curve has a unique profile which allows the detection of polymorphisms between DNA samples based on the differences in the melting temperature of PCR products, as low as in a single nucleotide such as SNPs (Simco, 2016; Muleo et al., 2009). The rate of fluorescent decrease is generally greatest near the melting temperature (Tm) of the PCR product. Therefore, a DNA sequence which has a unique combination of length, GC content and pattern of complementary bases will exhibit a unique melting curve or melting profile (Xanthopoulou et al., 2014).
|
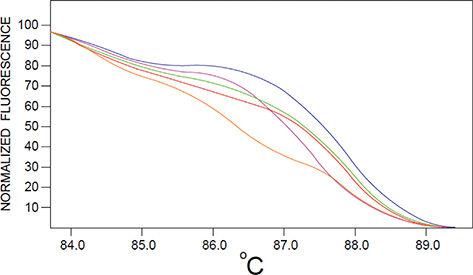 Figure 2. HRM normalized melting curve obtained from the diminishing fluorescent emission values plotted against increasing temperature. In this hypothetical example, 5 samples of extracted DNA from the olive oil of different olive cultivars were amplified by PCR using specific primers (targetting a SNP) and analyzed by HRM. The samples were distinguished by their unique melting curves (each sample corresponds to a different color line) [adapted from Song et al., 2016]. Figure 2. HRM normalized melting curve obtained from the diminishing fluorescent emission values plotted against increasing temperature. In this hypothetical example, 5 samples of extracted DNA from the olive oil of different olive cultivars were amplified by PCR using specific primers (targetting a SNP) and analyzed by HRM. The samples were distinguished by their unique melting curves (each sample corresponds to a different color line) [adapted from Song et al., 2016].
|
|
Small differences between individual melting curves are better visualized by the calculation of the HRM difference (or differential) plot provided by the HRM software (Pereira et al., 2018; Song et al., 2016). This plot is constructed by setting the melting profile of one of the known genotypes as a baseline reference and then by subtracting the normalized fluorescent data by this reference melting profile as shown in Figure 3.
|
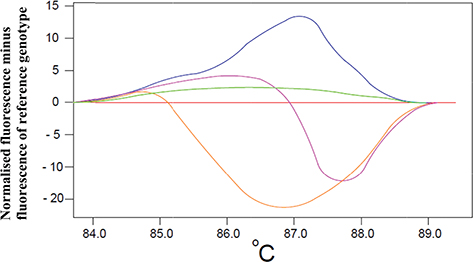 Figure 3. A HRM difference plot based on the analysis of 5 samples. One of the samples was used as a reference (reference genotype, the red line in this graph) and the fluorescence of the samples minus the fluorescence of the reference was calculated (vertical axis). This graph is used to better visualize the differences between individual melting curves [adapted from Song et al., 2016]. Figure 3. A HRM difference plot based on the analysis of 5 samples. One of the samples was used as a reference (reference genotype, the red line in this graph) and the fluorescence of the samples minus the fluorescence of the reference was calculated (vertical axis). This graph is used to better visualize the differences between individual melting curves [adapted from Song et al., 2016].
|
|
In general, the melting curve analysis software defines the Tm of a PCR product as the inflection point of the melting curve. To visualize the Tm more clearly, the negative first derivatives (−dF/dT) are often plotted, making the Tm of the products appear as peaks, as seen in Figure 4.
|
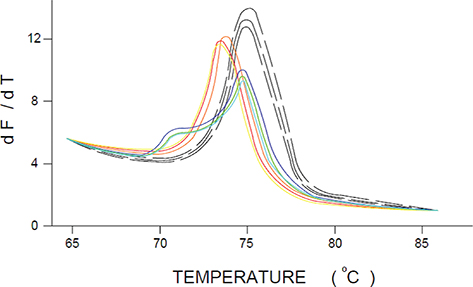 Figure 4. HRM derivative plot. This plot is used to visualize the melting temperatures Tm of the samples analyzed more clearly (Tm = the temperature at which 50% of the DNA is single stranded). The plot uses the negative first derivatives of fluorescence over temperature (−dF/dT). The Tms of the products appear as peaks [adapted from Liew et al., 2004]. Figure 4. HRM derivative plot. This plot is used to visualize the melting temperatures Tm of the samples analyzed more clearly (Tm = the temperature at which 50% of the DNA is single stranded). The plot uses the negative first derivatives of fluorescence over temperature (−dF/dT). The Tms of the products appear as peaks [adapted from Liew et al., 2004].
|
|
In the case of diploid organisms (whose genome has two copies of each chromosome) such as Olea plants, two alleles (variant forms of the same gene or genetic locus) may differ in only one base and can be used as molecular markers. Individual organisms can be either homozygous for an allele (having two copies of exactly the same sequence) or heterozygous (each copy has a slight difference e.g. one different base) (Nature, 2019).
In a heterozygote (assuming that allele A1 and allele A2 differ in only one base and therefore have a single nucleotide polymorphism, SNP), after PCR amplification of the selected target sequence where the SNP is located, four duplexes are formed: two homoduplexes (allele A1/complement of allele A1 and allele A2/complement of allele A2) and two heteroduplexes (allele A1/complement of allele A2 and allele A2/complement of allele A1). The melting curve analysis exhibits the melting profile of each duplex and reveals the presence of heteroduplexes (derived from heterozygotes) and homoduplexes (derived from homozygotes) because they have different melting curve shapes (Mehta et al., 2017; Liew et al., 2004; Gundry et al., 2003). High-resolution melting analysis can reliably detect the differences between homozygous samples which exhibit the sharpest melting transitions and heterozygous samples in which a SNP is present which shows broader transitions because the heteroduplexes formed are heterogeneous and lower-melting. In addition, heterozygotes that have different variations such as SNPs will produce different melting curve shapes because different heteroduplexes are formed (Reed and Whittwer, 2004). Therefore, the presence or absence of specific alleles in a sample provide information about the cultivars present or absent from that sample and these data could be used to justify fraud.
HRM analysis is also combined with an analysis named DNA barcoding which is based on a short, standardized fragment of the genome called “DNA barcode” (Hebert et al., 2003). The DNA barcode could be a locus of the chloroplast, mitochondrial or nuclear genome and is used to identify organisms at the species level (Pereira et al., 2018). The data derived from the analysis of DNA barcodes of genomes combined with the analysis of genetic markers such as microsatellites or Simple Sequence Repeat (SSR) and SNPs markers can be used for verification, identification and classification of cultivars, germplasm and species and also in the authentication of food (Lo and Shaw, 2018). HRM has some main advantages such as diminishing post PCR manipulation, closed-tube analysis, cost effectiveness, and the relatively rapid obtaining of results (Pereira et al., 2018) which have contributed to its wide application in plant research and, in particular, in the food sector as shown in Table 3.
Table 2. Applications of SNPs in the authentication of olive oil
| Year of publication |
Main Findings |
Reference |
| 2016 |
SNPs discriminated six among 13 mono-varietal (3 Greek and 3 Tunisian) olive oils using PCR-RFLP assay combined with capillary electrophoresis and detected olive oil admixtures down to a limit of 10%. |
Bazakos et al., 2016a |
| 2015 |
The first multiplex SNP genotyping assay for olive oil cultivar identification which was performed on a suspension of fluorescence-encoded microspheres. Three SNPs were adequate to discriminate five mono-varietal olive oils. |
Kalogianni et al., 2015 |
| 2015 |
Cleaved amplified polymorphic (CAP) DNA assay for SNPs which reside in restriction sites was used to discriminate 17 Turkish olive cultivars. The efficiency and limit of their approach for detecting olive oil admixtures was down to a limit of 20%. |
Uncu et al., 2015 |
| 2015 |
Five SNPs localized in four different genes were used to evaluate the associations between SNP and olive oil quality parameters in 16 Tunisian olive tree cultivars and their effectiveness in authenticity. Significant associations were found between SNPs and the content in polyunsaturated fatty acid (C18:2and C18:3), cholesterol rate, high and alternating productivity. |
Ben Ayed et al., 2015 |
| 2012 |
The SNP database of 10 Greek olive varieties was constructed and five SNPs, residing in restriction sites, were used in a PCR–RFLP capillary electrophoresis method. The five SNPs were adequate to discriminate nine out of the 10 using leaf DNA as template. |
Bazakos et al., 2012 |
Table 3. Applications of High Resolution Melting (HRM) in the authentication of olive oil
| Year of publication |
Main Findings |
Reference |
| 2018 |
HRM melting curve analysis based on nuclear microsatellites (gSSR) and plastid DNA (cpDNA) generated 14 HRM profiles sufficient to genotype all oil varieties present in oil samples. |
Gómez et al., 2018 |
| 2016 |
HRM curve profiles of DNA microsatellites of crude olive-pomace oil revealed the alteration of the declared varietal profile of a virgin olive oil |
Pasqualone et al., 2016b |
| 2015 |
HRM with 17 SSR markers was used for discriminating the olive cultivars used in the “Terra di Bari” PDO extra virgin olive oil showing different melting temperatures; and also for quantitative detection of the different levels of adulteration (10–20–30–40–50%) between the monovarietal oil Coratina and non-PDO Aeleh. |
Montemuro et al., 2015 |
| 2014 |
High-resolution melting (HRM) analysis, coupled with five microsatellite markers was used to characterize 47 O. europaea cultivars collected from the National Olive Tree Germplasm Collection established in Chania, Greece by generating a unique melting curve profile for each of the cultivars. Three microsatellite markers (DCA03, DCA09 and DCA17), which generated 29
HRM profiles, were sufficient to genotype all the olive cultivars studied. |
Xanthopoulou et al., 2014 |
| 2013 |
HRM analysis was performed with PCR primers for the rbcL gene in order to detect the presence of maize and sunflower oils in artificial mixtures of olive oils and particularly in olive maize oil mix and olive-sunflower oil mix in ratios (v/v) 50/50, 70/30, 80/20 and 90/10. The HRM results showed that both maize oil and sunflower oil could be detected down to 10% limit of
detection. |
Vietina et al., 2013 |
| 2013 |
The adulteration of olive (Olea europaea) oil with canola (Brassica napus) oil was analyzed with HRM and the results showed that the universal rbcL region is efficient enough to discriminate plant oil species and to detect the existence of 1% canola oil admixed into olive oil. |
Ganopoulos et al., 2013 |
| 2009 |
HRM analysis was performed on PCR products of a fragment of phytochrome A gene (phyA) from six cultivars of O. europaea L. The presence of SNPs was identified in all cultivars tested and enabled the grouping of the cultivars according to their genotype. |
Muleo et al., 2009 |
7. CONCLUSIONSTOP
There is an increasing global demand for high quality olive oil due to its established beneficial properties. Extra virgin olive oil produced in limited quantities by certain varieties and in specific geographical areas has premium prices that reflect its excellent quality and for that reason it is subjected to a high degree of adulteration. It is imperative to establish reliable and effective methods to assess varietal identification, genetic authenticity, traceability and adulteration. The use of DNA based methods aims to develop discriminatory tools that could provide accurate authentication since residual DNA is present in olive oil and its analysis (genotyping) could provide detailed information about its genetic origin (variety, subspecies, species) which in some cases is more accurate than biochemical methods that measure metabolites, which are strongly influenced by environmental factors. To this end, a number of specific biomarkers such as SSRs and SNPs found in the genome of Olea europaea have been used in many recent studies to detect the identity of DNA extracted from olive oil. High resolution melting (HRM), a method widely used in biomedical studies, enables a rapid, high-throughput identification of genetic variants in olive oil. In the future, the application of next-generation sequencing in food genomics and the development of quick protocols for on-site authentication will contribute substantially to the detection of genetic authenticity of olive oil and other valuable food products.
REFERENCESTOP
| ○ |
Abuzayed M, Frary A, Doganlar S. 2018. Genetic diversity of some Palestinian and Turkish olive (Olea europaea L.) germplasm determined with SSR markers. IUG J. Natural Stud. 26 (1), 10–17. |
| ○ |
Agrimonti C, Vietina M, Pafundo S, Marmiroli N. 2011. The use of food genomics to ensure the traceability of olive oil. Trends Food Sci. Technol. 22 (5), 237–244. https://doi.org/10.1016/j.tifs.2011.02.002 |
| ○ |
Alba V, Sabetta W, Blanco A, Pasqualone A, Montemurro C. 2009. Microsatellite markers to identify specific alleles in DNA extracted from monovarietal virgin olive oils. Eur. Food Res. Technol. 229 (3), 375–382. https://doi.org/10.1007/s00217-009-1062-8 |
| ○ |
Alonso-Rebollo A, Ramos-Gómez S, Busto MD, Ortega N. 2017. Development and optimization of an efficient qPCR system for olive authentication in edible oils. Food Chem. 232, 827–835. https://doi.org/10.1016/j.foodchem.2017.04.078 |
| ○ |
Álvarez-Rivera G, Cifuentes A, Puyana MC. 2018. Electrophoretic Technique: Capillary Zone Electrophoresis. In: Modern Techniques for Food Authentication (Second Edition, pp. 659–685). Academic Press. https://doi.org/10.1016/B978-0-12-814264-6.00016-5 |
| ○ |
Bazakos C, Dulger AO, Uncu AT, Spaniolas S, Spano T, Kalaitzis P. 2012. A SNP-based PCR-RFLP capillary electrophoresis analysis for the identification of the varietal origin of olive oils. Food Chem. 134 (4), 2411–2418. https://doi.org/10.1016/j.foodchem.2012.04.031 |
| ○ |
Bazakos C, Khanfir E, Aoun M, Spano T, Zein ZE, Chalak L, Riachy ME, Abou-Sleymane G, Ali SB, Grati Kammoun N, Kalaitzis P. 2016a. The potential of SNP-based PCR-RFLP capillary electrophoresis analysis to authenticate and detect admixtures of Mediterranean
olive oils. Electrophoresis 37 (13), 1881–1890. https://doi.org/10.1002/elps.201500537 |
| ○ |
Bazakos C, Spaniolas S, Kalaitzis P. 2016. DNA-Based Approaches for Traceability and Authentication of Olive Oil, Products from Olive Tree. D. Boskou and M.L. Clodoveo, Intech. Open, https://doi.org/10.5772/64494 |
| ○ |
Belaj A, Satovic Z, Cipriani G, Baldoni L, Testolin R, Rallo L, Trujillo I. 2003. Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theoret. App. Genet. 107, 736–744. https://doi.org/10.1007/s00122-003-1301-5 |
| ○ |
Ben Ayed R, Ben Hassen H, Ennouri K, Ben Marzoug R, Rebai A. 2016. OGDD (Olive Genetic Diversity Database): a microsatellite markers’ genotypes database of worldwide olive trees for cultivar identification and virgin olive oil traceability. Database. https://doi.org/10.1093/database/bav090 |
| ○ |
Ben-Ayed R, Kallel I, Hassen H, Rebai A. 2015. SNP marker analysis for validating the authenticity of Tunisian olive oil. J. Genet. 94 (1), 148–154. https://doi.org/10.1007/s12041-014-0461-1 |
| ○ |
Ben-Ayed R, Kamoun-Grati N, Rebai A. 2013. An overview of the authentication of olive tree and oil. Compr. Rev. Food Sci. Food Saf. 12 (2), 218–227. https://doi.org/10.1111/1541-4337.12003 |
| ○ |
Ben-Ayed R, Sans-Grout C, Moreau F, Grati-Kamoun N, Rebai A. 2014. Genetic similarity among Tunisian olive cultivars and two unknown feral olive trees estimated through SSR markers. Biochem. Genet. 52 (5–6), 258–268. https://doi.org/10.1007/s10528-014-9645-x |
| ○ |
Besnard G, Garcia-Verdugo C, De Casas RR, Treier UA, Galland N, Vargas P. 2008. Polyploidy in the olive complex (Olea europaea): evidence from flow cytometry and nuclear microsatellite analyses. Ann. Bot. 101 (1), 25–30. https://doi.org/10.1093/aob/mcm275 |
| ○ |
Besnard G, Rubio de Casas R, Christin PA, Vargas P. 2009. Phylogenetics of Olea (Oleaceae) based on plastid and nuclear ribosomal DNA sequences: tertiary climatic shifts and lineage differentiation times. Ann. Bot. 104 (1), 143–160. https://doi.org/10.1093/aob/mcp105 |
| ○ |
Bracci T, Busconi M, Fogher C, Sebastiani L. 2011. Overview on molecular studies in olive (Olea europaea L.): DNA markers application and first results in genome analysis. Plant Cell Rep. 30, 449–462. https://hdl.handle.net/10807/33192 |
| ○ |
Breton C, Claux D, Metton I, Skorski G, Bervillé A. 2004. Comparative study of methods for DNA preparation from olive oil samples to identify cultivar SSR alleles in commercial oil samples: possible forensic applications. J. Agric. Food Chem. 52 (3), 531–537. https://doi.org/10.1021/jf034588f |
| ○ |
Chiappetta A, Muto A, Muzzalupo R, Muzzalupo I. 2017. New rapid procedure for genetic characterization of Italian wild olive (Olea europaea) and traceability of virgin olive oils by means of SSR markers. Sci. Hortic. 226, 42–49. https://doi.org/10.1016/j.scienta.2017.08.022 |
| ○ |
Duran C, Appleby N, Edwards D, Batley J. 2009. Molecular genetic markers: discovery, applications, data storage and visualisation. Curr. Bioinform. 4 (1), 16–27. https://doi.org/10.2174/157489309787158198 |
| ○ |
European Council Regulation (EC) No 510/2006 of 20 March 2006 on the protection of geographical indications and designations of origin for agricultural products and foodstuffs, OJ L 93, 31.3.2006, p. 12–25. https://data.europa.eu/eli/reg/2006/510/oj |
| ○ |
FAOSTAT 2016, Food. “Agriculture Organization of the United Nations Statistics Division.” Economic and Social Development Department, Rome, Italy. http://faostat3.fao.org/home/E. Accessed 12 (2016). |
| ○ |
Galimberti A, De Mattia F, Losa A, Bruni I, Federici S, Casiraghi M, Martellos S, Labra M. 2013. DNA barcoding as a new tool for food traceability. Food Res. Int. 50 (1), 55–63. https://doi.org/10.1016/j.foodres.2012.09.036 |
| ○ |
Ganopoulos I, Bazakos C, Madesis P, Kalaitzis P, Tsaftaris A. 2013. Barcode DNA high resolution melting (Bar-HRM) analysis as a novel close-tubed and accurate tool for olive oil forensic use. J. Sci. Food Agric. 93 (9), 2281–2286. https://doi.org/10.1002/jsfa.6040 |
| ○ |
Gemas VJV, Almadanim MC, Tenreiro R, Martins A, Fevereiro P. 2004. Genetic diversity in the Olive tree (Olea europaea L. subs. europaea) cultivated in Portugal revealed by RAPD and ISSR markers. Genet. Resour. Crop Evol. 51, 501–11. https://doi.org/10.1023/B:GRES.0000024152.16021.40 |
| ○ |
Giménez MJ, Pistón F, Martín A, Atienza SG. 2010. Application of real-time PCR on the development of molecular markers and to evaluate critical aspects for olive oil authentication. Food Chem. 118 (2), 482–487. https://doi.org/10.1016/j.foodchem.2009.05.012 |
| ○ |
Gomes S, Breia R, Carvalho T, Carnide V, Martins-Lopes P. 2018. Microsatellite High-Resolution Melting (SSR-HRM) to Track Olive Genotypes: From Field to Olive Oil. J. Food Sci. 83 (10), 2415–2423. https://doi.org/10.1111/1750-3841.14333 |
| ○ |
Gundry CN, Vandersteen JG, Reed GH, Pryor RJ, Chen J, Wittwer CT. 2003. Amplicon melting analysis with labeled primers: a closed-tube method for differentiating homozygotes and heterozygotes. Clin. Chem. 49 (3), 396–406. https://doi.org/10.1373/49.3.396 |
| ○ |
Gupta M, Chyi YS, Romero-Severson J, Owen JL. 1994. Amplification of DNA markers from evolutionarily diverse genomes using single primers of simple-sequence repeats. Theoret. App. Genet. 89, 998–1006. https://doi.org/10.1007/BF00224530 |
| ○ |
Hebert PD, Cywinska A, Ball SL, Dewaard JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London. Series B: Biological Sciences. 270 (1512), 313–321. https://doi.org/10.1098/rspb.2002.2218 |
| ○ |
Heimler D, Cimato A, Pieroni A, Sani G, Tattini M. 2004. Seasonal trend of flavonoids, flavonoid glycosides and biflavonoids in ten olive cultivars. In II International Symposium on Olive Growing 356, 372–374. https://doi.org/10.17660/ActaHortic.1994.356.79 |
| ○ |
Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK. 2011. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83 (22), 8604–10. https://doi.org/10.1021/ac202028g |
| ○ |
Ipek M, Ipek A, Seker M, Gul MK. 2015. Association of SSR markers with contents of fatty acids in olive oil and genetic diversity analysis of an olive core collection. Genet. Mol. Res. 14 (1), 2241–2252. https://doi.org/10.4238/2015.March.27.10. |
| ○ |
Kalaitzis P, El-Zein Z. 2016. Olive oil authentication, traceability and adulteration detection using DNA-based approaches. Lipid Technol. 28 (10–11), 173–176. https://doi.org/10.1002/lite.201600048 |
| ○ |
Kalogianni DP, Bazakos C, Boutsika LM, Targem MB, Christopoulos TK, Kalaitzis P, Ioannou PC. 2015. Olive oil DNA fingerprinting by multiplex SNP genotyping on fluorescent microspheres. J. Agric. Food Chem. 63 (12), 3121–3128. https://doi.org/10.1021/jf5054657 |
| ○ |
Karki R, Pandya D, Elston RC, Ferlini C. 2015. Defining “mutation” and “polymorphism” in the era of personal genomics. BMC Med. Genomics 8 (37), 1–7. https://doi.org/10.1186/s12920-015-0115-z |
| ○ |
Lian DS, Zeng HS. 2017. Capillary Electrophoresis Based on Nucleic Acid Detection as Used in Food Analysis. Compr. Rev. Food Sci. Food Saf. 16 (6), 1281–1295. https://doi.org/10.1111/1541-4337.12297 |
| ○ |
Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E. Wittwer C. 2004. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 50 (7), 1156–1164. https://doi.org/10.1373/clinchem.2004.032136 |
| ○ |
Lo YT, Shaw PC. 2018. DNA-based techniques for authentication of processed food and food supplements. Food Chem. 240, 767–774. https://doi.org/10.1016/j.foodchem.2017.08.022 |
| ○ |
Madesis P, Ganopoulos I, Sakaridis I, Argiriou A, Tsaftaris A. 2014. Advances of DNA-based methods for tracing the botanical origin of food products. Food Res. Int. 60, 163–172. https://doi.org/10.1016/j.foodres.2013.10.042 |
| ○ |
Martinez-Gonzalez MA, Martin-Calvo N. 2016. Mediterranean diet and life expectancy; beyond olive oil, fruits, and vegetables. Curr. Opin. Clin. Nutr. Metab. Care 19 (6), 401–407. https://doi.org/10.1097/MCO.0000000000000316 |
| ○ |
Mehta B, Daniel R, McNevin D. 2017. HRM and SNaPshot as alternative forensic SNP genotyping methods. Forensic Sci. Med. Pathol. 13 (3), 293–301. https://doi.org/10.1007/s12024-017-9874-5 |
| ○ |
Montealegre C, Alegre MLM, García-Ruiz C. 2010. Traceability markers to the botanical origin in olive oils. J. Agric. Food Chem. 58 (1), 28–38. https://doi.org/10.1021/jf902619z |
| ○ |
Montemurro C, Miazzi MM, Pasqualone A, Fanelli V, Sabetta W, di Rienzo V. 2015. Traceability of PDO olive oil ‘Terra di Bari’ using high resolution melting. J. Chem. Article ID 496986, 7 pages. https://doi.org/10.1155/2015/496986 |
| ○ |
Montemurro C, Pasqualone A, Simeone R, Sabetta W, Blanco A. 2008. AFLP molecular markers to identify virgin olive oils from single Italian cultivars. Eur. Food Res. Technol. 226, 1439–44. https://doi.org/10.1007/s00217-007-0675-z |
| ○ |
Mousavi S, Mariotti R, Regni L, Nasini L, Bufacchi M, Pandolfi S, Baldoni L, Proietti P. 2017. The first molecular identification of an olive collection applying standard simple sequence repeats and novel expressed sequence tag markers. Front. Plant Sci. 8, 1283. https://doi.org/10.3389/fpls.2017.01283 |
| ○ |
Muleo R, Colao MC, Miano D, Cirilli M, Intrieri MC, Baldoni L, Rugini E. 2009. Mutation scanning and genotyping by high-resolution DNA melting analysis in olive germplasm. Genome 52 (3), 252–260. https://doi.org/10.1139/G09-002 |
| ○ |
Muzzalupo I, Perri E. 2002. Recovery and characterisation of DNA from virgin olive oil. Eur. Food Res. Technol. 214, 528–531. https://doi.org/10.1007/s00217-001-0482-x |
| ○ |
Muzzalupo I, Pellegrino M, Perri E. 2007. Detection of DNA in virgin olive oils extracted from destoned fruits. Eur. Food Res. Technol. 224 (4), 469–475. https://doi.org/10.1007/s00217-006-0340-y |
| ○ |
Muzzalupo I, Pisani F, Greco F, Chiappetta A. 2015. Direct DNA amplification from virgin olive oil for traceability and authenticity. Eur. Food Res. Technol. 241 (1), 151–155. https://doi.org/10.1007/s00217-015-2455-5 |
| ○ |
Nature 2019, Nature.com, Springer Nature Publishing AG, https://www.nature.com/subjects/genetic-markers. Nat. Educ. 2014, https://www.nature.com/scitable/definition/allele-48 |
| ○ |
NCBI National Center for Biotechnology Information, genome https://www.ncbi.nlm.nih.gov/genome/10724 |
| ○ |
NCI 2019, National Cancer Institute Dictionary of Genetics Terms, US Department of Health and Human Services, National Institutes of Health, https://www.cancer.gov/publications/dictionaries/genetics-dictionary/def/genetic-marker |
| ○ |
Pafundo S, Agrimonti C, Marmiroli N. 2005. Traceability of plant contribution in olive oil by amplified fragment length polymorphisms. J. Agric. Food Chem. 5, 6995–7002. https://doi.org/10.1021/jf050775x |
| ○ |
Parkinson L, Cicerale S. 2016. The health benefiting mechanisms of virgin olive oil phenolic compounds. Molecules 21 (12), 1734. https://doi.org/10.3390/molecules21121734 |
| ○ |
Pasqualone A, Caponio F, Blanco A. 2001. Inter-simple sequence repeat DNA markers for identification of drupes from different Olea europaea L. cultivars. Eur. Food Res. Technol. 213, 240–3. https://doi.org/10.1007/s002170100367 |
| ○ |
Pasqualone A, Di Rienzo V, Blanco A, Summo C, Caponio F, Montemurro C. 2012. Characterization of virgin olive oil from Leucocarpa cultivar by chemical and DNA analysis. Food Res. Int. 47 (2), 188−193. https://doi.org/10.1016/j.foodres.2011.05.008 |
| ○ |
Pasqualone A, Di Rienzo V, Sabetta W, Fanelli V, Summo C, Paradiso VM, Montemurro C, Caponio F. 2016b. Chemical and molecular characterization of crude oil obtained by olive-pomace recentrifugation. J. Chem. 2016b, Article ID 4347207, 7 pg. https://doi.org/10.1155/2016/4347207 |
| ○ |
Pasqualone A, Montemurro C, di Rienzo V, Summo C, Paradiso VM, Caponio F. 2016. Evolution and perspectives of cultivar identification and traceability from tree to oil and table olives by means of DNA markers. J. Sci. Food Agric. 96 (11), 3642–3657. https://doi.org/10.1002/jsfa.7711 |
| ○ |
Pasqualone A, Montemurro C, Summo C, Sabetta W, Caponio F, Blanco A. 2007. Effectiveness of microsatellite DNA markers in checking the identity of Protected Designation of Origin extra virgin olive oil. J. Agric. Food. Chem. 55 (10), 3857–3862. https://doi.org/10.1021/jf063708r |
| ○ |
Pereira L, Gomes S, Barrias S, Fernandes R, Martins-Lopes P. 2018. Applying high-resolution melting (HRM) technology to olive oil and wine authenticity. Food Res. Int. 103, 170–181. https://doi.org/10.1016/j.foodres.2017.10.026 |
| ○ |
Primrose S, Woolfe M, Rollinson S. 2010. Food forensics: methods for determining the authenticity of foodstuffs. Trends Food Sci. Technol. 21 (12), 582–590. https://doi.org/10.1016/j.tifs.2010.09.006 |
| ○ |
Raieta K, Muccillo L, Colantuoni V. 2015. A novel reliable method of DNA extraction from olive oil suitable for molecular traceability. Food Chem. 172, 596–602. https://doi.org/10.1016/j.foodchem.2014.09.101 |
| ○ |
Ramos-Gómez S, Busto MD, Albillos SM, Ortega N. 2016. Novel qPCR systems for olive (Olea europaea L.) authentication in oils and food. Food Chem. 194, 447–454. https://doi.org/10.1016/j.foodchem.2015.08.036 |
| ○ |
Ramos-Gómez S, Busto MD, Perez-Mateos M, Ortega N. 2014. Development of a method to recovery and amplification DNA by real-time PCR from commercial vegetable oils. Food Chem. 158, 374–383. https://doi.org/10.1016/j.foodchem.2014.02.142 |
| ○ |
Reed GH, Wittwer CT. 2004. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin. Chem. 50 (10), 1748–1754. https://doi.org/10.1373/clinchem.2003.029751 |
| ○ |
Regulation, E. U. (2012). 432/2012. Commission Regulation (EU) No 432/2012 of 16 May 2012 establishing a list of permitted health claims made on foods, other than those referring to the reduction of disease risk and to children’s development and health. Official Journal of the European Union L, 136, 1–40. |
| ○ |
Scollo F, Egea LA, Gentile A, La Malfa S, Dorado G, Hernandez P. 2016. Absolute quantification of olive oil DNA by droplet digital-PCR (ddPCR): comparison of isolation and amplification methodologies. Food Chem. 213, 388–394. https://doi.org/10.1016/j.foodchem.2016.06.086 |
| ○ |
Sebastiani L, Busconi M. 2017. Recent developments in olive (Olea europaea L.) genetics and genomics: applications in taxonomy, varietal identification, traceability and breeding. Plant Cell. Rep. 36 (9), 1345–1360. https://doi.org/10.1007/s00299-017-2145-9 |
| ○ |
Sefc KM, Lopes MS, Mendonça D, Dos Santos MR, Machado MLDC, Machado ADC. 2000. Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Mol. Ecol. 9 (8), 1171–1173. https://doi.org/10.1046/j.1365-294x.2000.00954.x |
| ○ |
Simko I. 2016. High-resolution DNA melting analysis in plant research. Trends Plant Sci. 21 (6), 528–537. https://doi.org/10.1016/j.tplants.2016.01.004 |
| ○ |
Song M, Li J, Xiong C, Liu H, Liang J. 2016. Applying high-resolution melting (HRM) technology to identify five commonly used Artemisia species. Sci. Rep. 6, 34133. https://doi.org/10.1038/srep34133 |
| ○ |
Spaniolas S, Bazakos C, Awad M, Kalaitzis P. 2008. Exploitation of the chloroplast trnL (UAA) intron polymorphisms for the authentication of plant oils by means of a Lab-on-a-Chip capillary electrophoresis system. J. Agric. Food Chem. 56 (16), 6886–6891. https://doi.org/10.1021/jf8008926 |
| ○ |
Spaniolas S, Bazakos C, Spano T, Zoghby C, Kalaitzis P. 2010. The potential of plastid trnL (UAA) intron polymorphisms for the identification of the botanical origin of plant oils. Food Chem. 122 (3), 850–856. https://doi.org/10.1016/j.foodchem.2010.02.039 |
| ○ |
Syvänen AC. 2001. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2 (12), 930–942. https://doi.org/10.1038/35103535 |
| ○ |
Taylor SC, Laperriere G, Germain H. 2017. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Scientific Reports 7 (1), 2409. https://doi.org/10.1038/s41598-017-02217-x |
| ○ |
Terzopoulos PJ, Kolano B, Bebeli PJ, Kaltsikes PJ, Metzidakis I. 2005. Identification of Olea europaea L. cultivars using inter-simple sequence repeat markers. Sci. Hort. 105, 45–51. https://doi.org/10.1016/j.scienta.2005.01.011 |
| ○ |
Trichopoulou A, Martínez-González MA, Tong TY, Forouhi NG, Khandelwal S, Prabhakaran D, Mozaffarian D, de Lorgeril M. 2014. Definitions and potential health benefits of the Mediterranean diet: views from experts around the world. BMC Med. 12 (1), 112. https://doi.org/10.1186/1741-7015-12-112 |
| ○ |
Trujillo I, Ojeda MA, Urdiroz NM, Potter D, Barranco D, Rallo L, Diez CM. 2014. Identification of the Worldwide Olive Germplasm Bank of Córdoba (Spain) using SSR and morphological markers. Tree Genet. Genomes 10 (1), 141–155. https://doi.org/10.1007/s11295-013-0671-3 |
| ○ |
Uncu AT, Frary A, Doganlar S. 2015. Cultivar origin and admixture detection in Turkish olive oils by SNP-based CAPS assays. J. Agric. Food Chem. 63 (8), 2284–2295. https://doi.org/10.1021/acs.jafc.5b00090 |
| ○ |
Uncu AT, Uncu AO, Frary A, Doganlar S. 2017. Barcode DNA length polymorphisms vs fatty acid profiling for adulteration detection in olive oil. Food Chem. 221, 1026–1033. https://doi.org/10.1016/j.foodchem.2016.11.059 |
| ○ |
Vieira MLC, Santini L, Diniz AL, Munhoz CDF. 2016. Microsatellite markers: what they mean and why they are so useful. Genet. Mol. Biol. 39 (3), 312–328. https://doi.org/10.1590/1678-4685-GMB-2016-0027 |
| ○ |
Vietina M, Agrimonti C, Marmiroli M, Bonas U, Marmiroli N. 2011. Applicability of SSR markers to the traceability of monovarietal olive oils. J. Sci. Food Agric. 91 (8), 1381–1391. https://doi.org/10.1002/jsfa.4317 |
| ○ |
Vietina M, Agrimonti C, Marmiroli N. 2013. Detection of plant oil DNA using high resolution melting (HRM) post PCR analysis: a tool for disclosure of olive oil adulteration. Food Chem. 141 (4), 3820–3826. https://doi.org/10.1016/j.foodchem.2013.06.075 |
| ○ |
Xanthopoulou A, Ganopoulos I, Koubouris G, Tsaftaris A, Sergendani C, Kalivas A, Madesis P. 2014. Microsatellite high-resolution melting (SSR-HRM) analysis for genotyping and molecular characterization of an Olea europaea germplasm collection. Plant Genet. Resour. Newsl. 12 (3), 273–277. https://doi.org/10.1017/S147926211400001X |
| ○ |
Zhan MM, Cheng ZZ, Su GC, Wang AY, Chen HP, Yang ZS, Shan Z, Huang QM. 2015. Genetic relationships analysis of olive cultivars grown in China. Genet. Mol. Res. 2, 5958–5969. https://doi.org/10.4238/2015.June.1.13 |
Figure 1. Simplified diagram of DNA analysis of olive oil: a) DNA extraction of the olive oil, b) selection of specific genetic markers as target DNA which will enable discrimination among cultivars, establish genetic authenticity, or detect adulteration with different species. These genetic markers are polymorphic DNA sequences located in either nuclear, chloroplast or mitochondrial DNA and could be SNPs, SSRs, RAPD, ISSR, AFLP, etc. c) application of PCR with the appropriate primers which amplify the target DNA, d) genotyping or visualization of amplified target sequences by methods that can distinguish differences, even in one base, among DNA sequences.